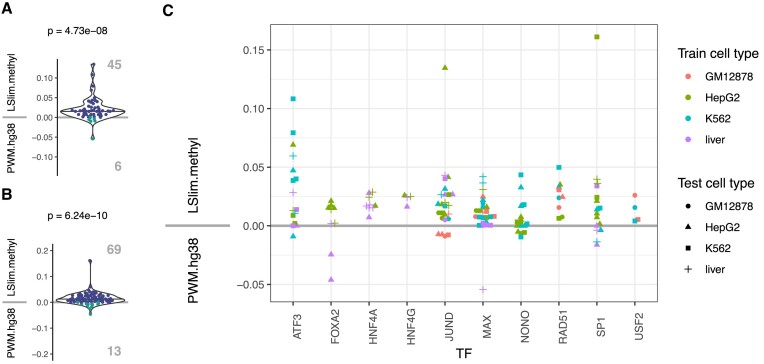
Figure 9.
Comparison of the genome-wide prediction performance of PWM models learned on the original hg38 genome (PWM.hg38) and methylation-aware LSlim models (LSlim.methyl) within the Catchitt framework. (A) Pairwise comparison of models across all 10 TFs in the within cell type setting. LSlim.methyl performs better than PWM.hg38 for 45 test data sets, while PWM.hg38 performs better than LSlim.methyl for 6 test data sets. (B) Pairwise comparison of models across all 10 TFs in the across cell type setting. LSlim.methyl performs better than PWM.hg38 for 69 test data sets, while PWM.hg38 performs better than LSlim.methyl for 13 test data sets. (C) Comparison of performance per TF, where the training cell type is encoded by colour and the test cell type is encoded by shape. If multiple data sets are present per TF and cell type, all combinations of data sets are considered.