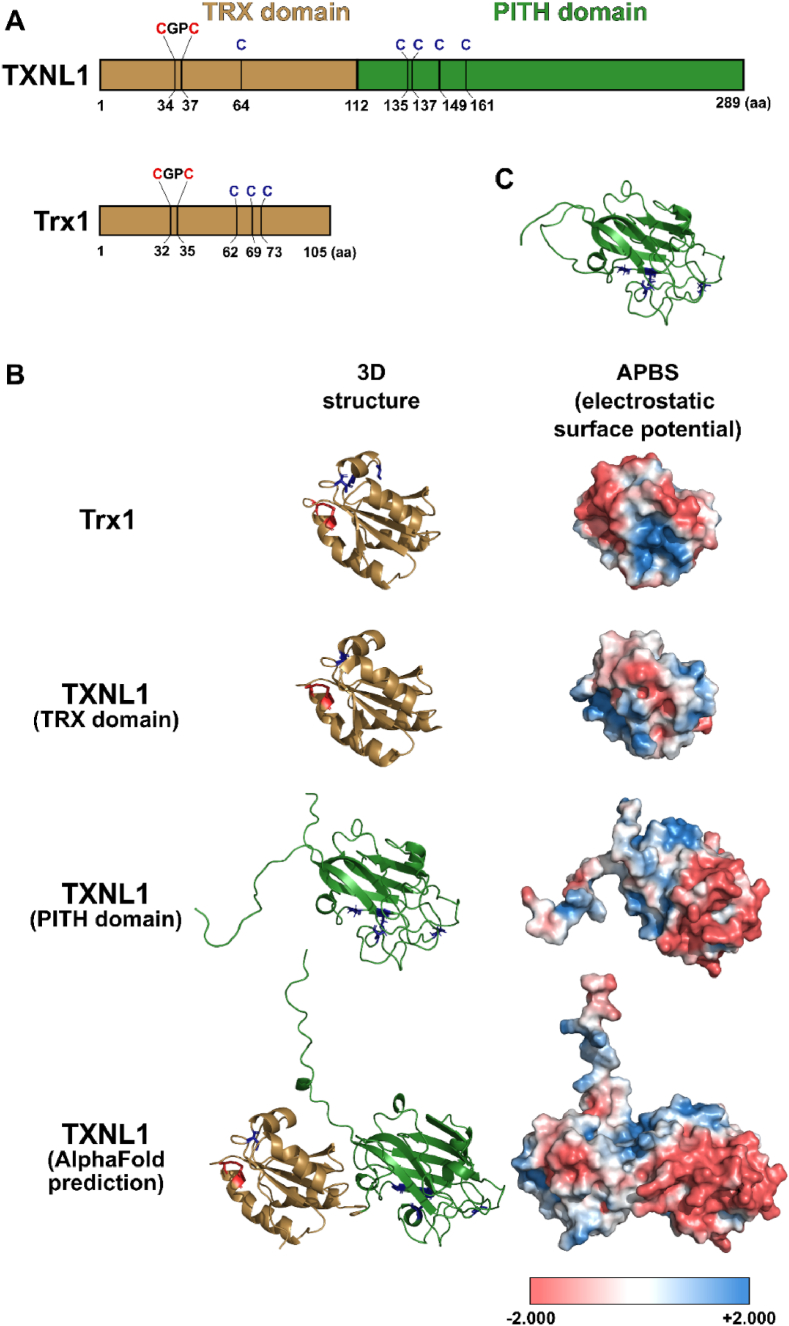
Fig. 1.
Domain organization and predicted surface features of human TXNL1 and Trx1. (A) Schematic domain organization of TXNL1 and Trx1 with their conserved active site sequence motifs (CGPC), typical of Trx proteins. Active site cysteines are highlighted and numbered in the schemes of both proteins (red), as are the additional Cys residues in both proteins (blue). (B) X-ray structures of Trx1 (PDB-structure: 1ERU) and the isolated Trx-domain of TXNL1 (PDB-structure: 1GH2), as well as the NMR-structure of the isolated PITH domain of TXNL1 (PDB-structure: 1WWY) together with the full-length structure of TXNL1 as predicted by AlphaFold (AF-O43396-F1), shown in the same orientation and scale using PyMol software and with the Cys residues shown as balls and sticks and color marked as in (A). In the right column of panel (B), the predicted electrostatic surface potentials of the proteins or domains are shown in the same orientation as in the left column, as visualized using the APBS plugin of PyMol; blue color is positive surface potential and red is negative. (C) Video (available in the online version) demonstrating the high flexibility of the C-terminal tail of the PITH domain of TXNL1 as found in prior NMR analyses (PDB structure: 1WWY). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)