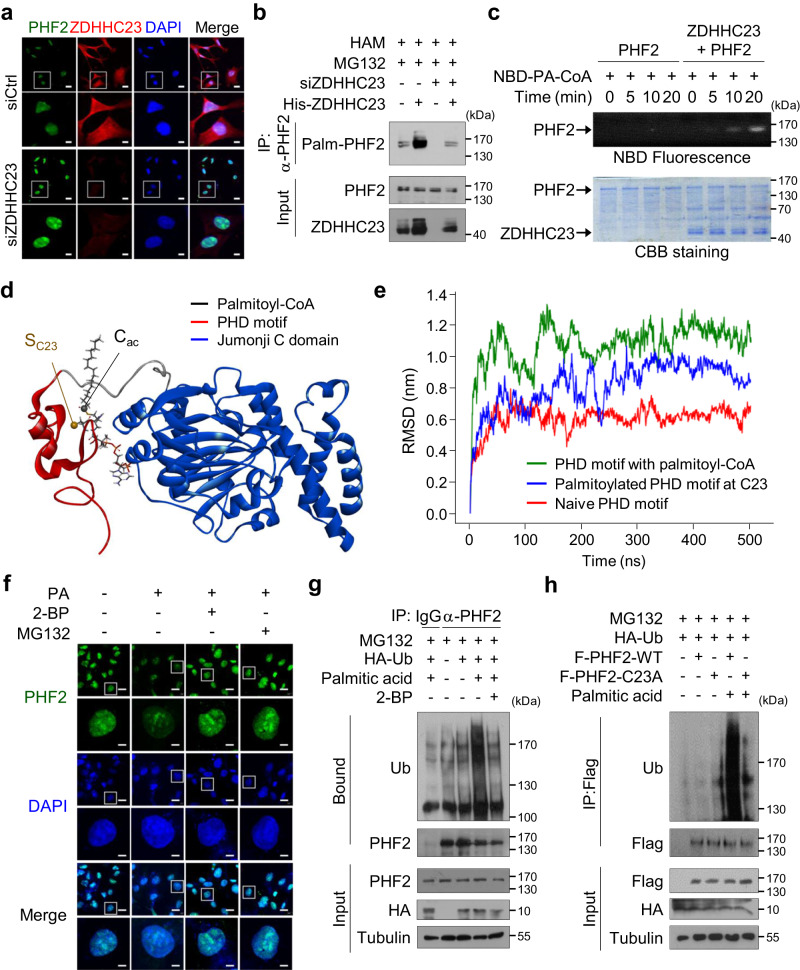
Fig. 2. ZDHHC23-mediated PHF2 palmitoylation at cysteine 23 enhances its ubiquitination.
a Hep3B cells were transfected with siZDHHC23 and immunofluorescence analysis was performed using indicated antibodies. Scale bar = 20 (low magnification) and 6 µm (high magnification). b HepG2 cells were transfected with siZDHHC23 or His-ZDHHC23, and treated with MG132. After immunoprecipitation with an anti-PHF2 antibody, the palmitoylated PHF2 was detected using ABE assay. n = 3 independent experiments. c An in vitro palmitoylation assay was performed using purified PHF2 and ZDHHC23 proteins. PHF2 palmitoylation was detected using a fluorescence-labeled palmitoyl-CoA analog. The proteins used in the assay were stained using Coomassie Brilliant Blue staining. n = 3 independent experiments. NBD-PA-CoA: (N-[(7-nitro-2-1,3-benzoxadiazol-4-yl)-methyl] amino) palmitoyl-coenzyme A. d 3D structure of PHF2 when the palmitoyl-CoA is in close proximity to C23 of the PHD domain (d = 5 Å). Red and blue ribbons represent the plant homeodomain (PHD) and Jumonji C domain, respectively, and sticks represent palmitoyl-CoA. e The time evolution of root mean square deviation (RMSD) for the PHD motif in the presence of palmitoyl-CoA (d = 5 Å), the PHD motif in the absence of palmitoyl-CoA, and the palmitoylated PHD domain in target to those of native particulate methane monooxygenase (pMMO) as a reference structure. f Hep3B cells were pre-treated with 2-BP and incubated with PA, followed by treatment with MG132. The level of PHF2 protein was determined using immunofluorescence. Scale bar = 20 (low magnification) and 5 µm (high magnification). g HepG2 cells were transfected with the indicated plasmids and treated with 2-BP and PA. After treatment of MG132 for 8 h, cells were immunoprecipitated using an anti-PHF2 antibody and subjected to immunoblotting. n = 3 independent experiments. h Flag-PHF2-WT or Flag-PHF2-C23A transfected cells were treated with PA and MG132. Immunoprecipitation was performed using Flag affinity beads. n = 3 independent experiments. F: Flag. Source data are provided as a Source Data file.