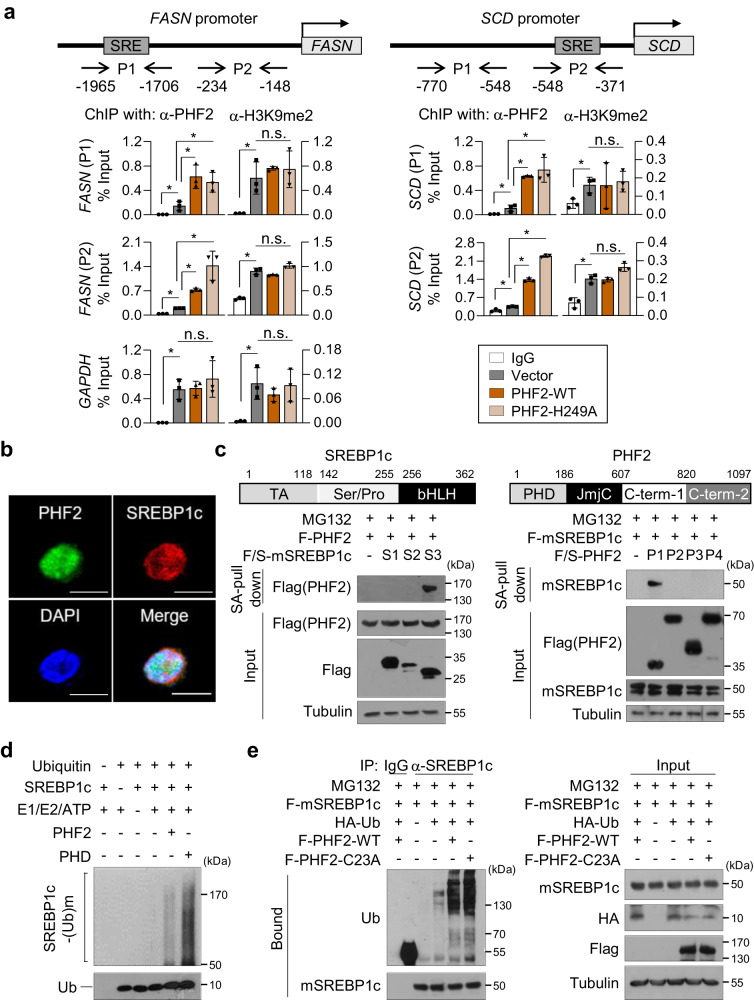
Fig. 6. PHF2 destroys SREBP1c as an E3 ubiquitin ligase.
a HepG2 cells were transfected with the indicated plasmids. Cell lysates were subjected to ChIP-qPCR assays using anti-H3K9me2 or anti-PHF2 antibodies at the FASN or SCD promoters. GAPDH was used as a negative control; mean ± SD (n = 3 independent experiments); *P < 0.05. Statistical analyses were based on a two-tailed unpaired t-test. The exact p-values are shown in Supplementary Data 2. P1, P2: promoter 1, 2; H3K9me2: histone H3 lysine 9 dimethylation. b Immunofluorescence analysis was performed using the indicated antibodies in Hep3B cells. n = 3 independent experiments. Scale bar = 10 µm. c (Top) Schematic diagram of the segments of SREBP1c or PHF2. The indicated plasmids were transfected into HepG2 cells (Bottom, left) or 293 T cells (Bottom, right). Then, cells were subjected to immunoprecipitation and immunoblotting after MG132 treatment for 8 h. Western blotting using the indicated antibodies evaluated the purified proteins using SA affinity beads. n = 3 independent experiments. TA: a transcription-activation domain; Ser/Pro: a serine- and proline-rich region; bHLH: a basic helix-loop-helix domain; PHD: a plant homeodomain; JmjC: a Jumonji C domain; C-term: C-terminus. d An in vitro ubiquitination assay was performed. The enzymatic reaction was stopped by adding a sample buffer and proteins were analyzed by western blotting. n = 3 independent experiments. e HepG2 cells were transfected with the indicated plasmids and then incubated with MG132 for 8 h. Cell lysates were pull-downed with the anti-SREBP1 antibody, and the pull-downed proteins were analyzed by western blotting using the indicated antibodies. n = 3 independent experiments. Source data are provided as a Source Data file.