Abstract
Purpose:
Prognostic and predictive biomarkers to cyclin-dependent kinases 4 and 6 inhibitors are lacking. Circulating tumor DNA (ctDNA) can be used to profile these patients and dynamic changes in ctDNA could be an early predictor of treatment efficacy. Here, we conducted plasma ctDNA profiling in patients from the PEARL trial comparing palbociclib+fulvestrant versus capecitabine to investigate associations between baseline genomic landscape and on-treatment ctDNA dynamics with treatment efficacy.
Experimental Design:
Correlative blood samples were collected at baseline [cycle 1-day 1 (C1D1)] and prior to treatment [cycle 1-day 15 (C1D15)]. Plasma ctDNA was sequenced with a custom error-corrected capture panel, with both univariate and multivariate Cox models used for treatment efficacy associations. A prespecified methodology measuring ctDNA changes in clonal mutations between C1D1 and C1D15 was used for the on-treatment ctDNA dynamic model.
Results:
201 patients were profiled at baseline, with ctDNA detection associated with worse progression-free survival (PFS)/overall survival (OS). Detectable TP53 mutation showed worse PFS and OS in both treatment arms, even after restricting population to baseline ctDNA detection. ESR1 mutations were associated with worse OS overall, which was lost when restricting population to baseline ctDNA detection. PIK3CA mutations confer worse OS only to patients on the palbociclib+fulvestrant treatment arm. ctDNA dynamics analysis (n = 120) showed higher ctDNA suppression in the capecitabine arm. Patients without ctDNA suppression showed worse PFS in both treatment arms.
Conclusions:
We show impaired survival irrespective of endocrine or chemotherapy-based treatments for patients with hormone receptor–positive/HER2-negative metastatic breast cancer harboring plasma TP53 mutations. Early ctDNA suppression may provide treatment efficacy predictions. Further validation to fully demonstrate clinical utility of ctDNA dynamics is warranted.
Translational Relevance.
Our results show that in patients with hormone receptor–positive/HER2-negative metastatic breast cancer, circulating tumor DNA (ctDNA) detection, likely reflecting higher tumor burden, is a poor prognostic biomarker. In addition, detection of plasma TP53 mutations confer impaired survival irrespective of endocrine or chemotherapy-based treatments, even after correction for known relevant clinicopathologic variables and ctDNA detection itself. We suggest this observation should be added to prognostic models and taken into account when designing therapeutic strategies for this population. Other plasma mutations in relevant genes like PIK3CA and in particular ESR1, might be associated with worse long-term outcomes. Our study also shows early ctDNA suppression can select optimal responders in both treatment arms and adds evidence that ctDNA dynamics might be a clinically useful tool for treatment efficacy predictions, although further validation would be required.
Introduction
Cyclin-dependent kinases 4 and 6 inhibitors (CDK4/6i) in combination with endocrine therapy (ET) is the mainstay treatment for patients with hormone receptor–positive (HR+), HER2-negative (HER2−) metastatic breast cancer (MBC), both in endocrine-sensitive (1–3) and resistant scenarios (4–6). However, efficacy of this combination was not originally tested in comparison with chemotherapy, an optional standard treatment in pretreated patients. The GEICAM/2013–02 PEARL study (ClinTrials.gov reference NCT02028507) was a multicenter randomized phase III clinical trial that enrolled patients with aromatase inhibitor (AI)-resistant HR+/HER2− MBC. Overall, the trial concluded there was no superiority of palbociclib plus ET over capecitabine in AI-resistant patients, although a more favorable safety profile was found for patients treated with CDK4/6i compared with capecitabine (7), reinforcing the evidence that CDK4/6i should be considered upfront for the treatment of HR+/HER2− MBC patients, but prognostic and predictive biomarkers to better select patients likely to benefit from these treatments is an area of intense research.
Plasma circulating tumor DNA (ctDNA) is shed into circulation by tumor cells in small fragments (typically around 143–145 bp), thought mainly to derive from tumor necrosis and apoptosis mechanisms (8). In theory, the pool of plasma ctDNA should better capture the temporal and spatial heterogeneity of the tumor across a patient cancer history, serving as an ideal genetic source for biomarkers.
Work using ctDNA assays in plasma from patients randomized to palbociclib or placebo in addition to fulvestrant in the PALOMA-3 trial showed high ctDNA fraction; TP53 mutations and FGFR1 amplifications were associated with worse outcome with no interaction between treatments (9). Mutations in other functionally important genes in MBC like PIK3CA and ESR1 have not shown clinical significance for treatment decisions on CDK4/6i in patients with similar clinical situation to those enrolled in PEARL (9, 10).
Dynamic changes in ctDNA as a result of therapeutic pressure have been proposed as a tool for early treatment efficacy predictions, and work conducted in patients treated with CDK4/6i combinations had previously reported high on-treatment ctDNA as an adverse biomarker predicting worse progression-free survival (PFS; refs. 11–14), although different technologies and ctDNA change calculations have been used, preventing standardization of a methodology. Exploratory work in limited cohorts of MBC patients treated with different targeted therapies sought to compare various strategies to optimize a cutoff for predictions, proposing an optimal method to further explore in larger cohorts (15).
In this work, we conducted plasma ctDNA profiling in patients treated in cohort 2 of the PEARL trial to investigate associations between baseline genomic landscape and on-treatment plasma ctDNA dynamics with efficacy.
Materials and Methods
Clinical trial and patients
The GEICAM/2013–02 (PEARL) clinical trial was a phase III, multicenter, international, open-label, randomized study that enrolled two successive cohorts of patients with AI-resistant HR+/HER2− MBC. Cohort 1 included 296 patients randomized to palbociclib plus exemestane versus capecitabine. Upon discovery of ESR1 mutations conferring resistance to further AI (16), cohort 2 was added and included 305 patients randomized to palbociclib plus fulvestrant versus capecitabine, as described before (7). Eligible patients for both cohorts had to be postmenopausal women with AI-resistant HR+ HER2− MBC, defined as recurrence while on or within 12 months after the end of adjuvant treatment or progression while on or within 1 month after the end of treatment for advanced disease. Patients must have an Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1 and measurable disease according to the RECIST (version 1.1) or at least one lytic or mixed bone lesion. One prior line of chemotherapy for MBC was permitted. Exclusion criteria included prior treatment with CDK4/6i, mTOR or PI3K inhibitors, capecitabine, or visceral crisis. The study was conducted in compliance with the Declaration of Helsinki. GEICAM/2013–02 (PEARL) clinical trial (NCT02028507) research protocol was approved by every site's institutional review board and every country's regulatory agency. All patients signed written informed consents.
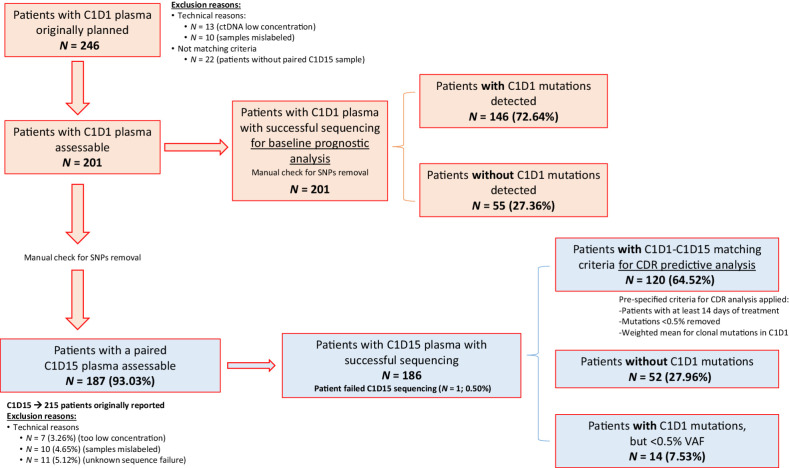
For this work, we focused on patients from cohort 2. A schematic view of samples used is given in Fig. 1. Blood samples were collected for ctDNA analysis from -7 days to cycle 1-day 1 (C1D1) for baseline prognostic analysis and cycle 1-day 15 (C1D15) when available. A total of 246 different patients with C1D1 were considered. 13 patients had DNA concentrations too low (under 0.10 ng/μL) so were discarded. From remaining 233 patients, 22 did not have an additional follow-up sample at C1D15 to have a paired analysis, so were also discarded. From remaining 211 patients, 10 failed as result of mislabeling during aliquoting, resulting in a total of 201 final different patients included in the analysis, all with a baseline and at least 1 follow-up sample.
Figure 1.
CONSORT diagram of the study population and samples. Blood samples were collected for ctDNA analysis from -7 days to C1D1 for baseline prognostic analysis and C1D15 when available. A total of 246 different patients with C1D1 were considered. 13 patients had DNA concentrations too low (under 0.10 ng/μL) so were excluded. From remaining 233, 22 did not have an additional follow-up sample at C1D15 to have a paired analysis, so were also excluded. From remaining 211, 10 samples were excluded as result of mislabeling during aliquoting, resulting in a total of 201 final different patients included in the analysis, all with a baseline and at least 1 follow-up sample.
Sample processing
Correlative blood samples were collected from each PEARL study cohort 2 patient at two extraction times: baseline (C1D1) and prior to treatment on C1D15 (±3 days).
For the ctDNA analysis, three 10-mL blood samples were collected directly in Cell-Free DNA BCT tubes (Streck Corporate) by venepuncture. Blood was mixed with the preservative in the BCT by gentle inversion and plasma processed within 2 hours after extraction. The procedure, specified in the PEARL study Sample Management Manual, consists of a single 20-minute centrifugation step at 1,600 g and immediate transfer of plasma into labelled cryovials and storage at −80°C.
Circulating plasma DNA was extracted from 4 mL of plasma sample using the MagMAX Cell-Free DNA Isolation Kit (Thermo Scientific, catalog no. A29319) on a KingFisher Flex Purification System as per the manufacturer's instructions. Extracted cell-free DNA was stored at −20°C until further use.
ctDNA sequencing
Plasma DNA was captured for targeted sequencing using the Nonacus Cell3 Target Cell-Free DNA Target Enrichment System protocol (Birmingham, UK), with 1.8 to 50.1ng (average of 23.5 ng) plasma DNA with no additional fragmentation. The DNA was tagged using dual index Unique Molecular Identifier (UMI) adapters at a concentration of 15 μmol/L for DNA input of >20 to 50 ng or 7.5 μmol/L for ≤20 ng input prior to six or eight cycles of PCR respectively. Libraries were then pooled by equal mass and hybridized overnight with a custom capture panel designed to target the coding regions of the 21 most commonly mutated genes in breast cancer (Supplementary Table S1). Post capture hybridisation, libraries were cleaned and amplified using 13 cycles of PCR according to the manufacturer's instructions. Amplified libraries were sequenced on an Illumina NovaSeq6000 (Illumina, San Diego, CA) in the Centre for Molecular Pathology at The Royal Marsden Hospital NHS Foundation Trust (London, United Kingdom), with 100 bp paired-end reads and v1.5 chemistry according to the manufacturer's instructions. 4 Gb of sequence data was allocated for plasma DNA samples, resulting in mean unique deduplicated UMI consensus depth of 1978x and 1620x for C1D1 and C1D15 samples, respectively.
Data analysis
A bespoke pipeline (MDIMSv4) developed in-house by the Translational Research Group at The Royal Marsden Hospital NHS Foundation Trust was used to analyze the ctDNA data.
Briefly, base calls were demultiplexed using bcl2fastq2 (v2.20.0). Reads had adapters removed with Picard (v2.23.8) and were aligned to the reference genome (GRCh37/Hg19) using BWA-MEM (v0.7.17). fgbio (v1.3.0) was used to collapse reads from the same original molecule using UMIs. UMI families were defined as containing ≥3 molecules with the same UMI. UMI families containing <3 reads were excluded from further analysis. Overlapping forward and reverse reads were hard clipped with ClipBam (fgbio). Picard was used for quality control (QC). BAM files were base recalibrated using GATK (v4.1.9.0) and finally variant calling was performed with VarDict (v1.8.2) without germline subtraction. Likely germline calls based on a Variant Population Frequency in general population in the Genome Aggregation Database (https://gnomad.broadinstitute.org/) >0.0001 were removed from the dataset. Analysis and filtering of the data using custom R scripts of potential germline mutations based on variant allele frequency (VAF) and oncogenic status evaluated with restriction to oncogenic/likely oncogenic variants. Remaining variants were manually checked using Integrative Genome Viewer (IGV; Broad Institute and the Regents of the University of California). Variant calling has been validated to a sensitivity and specificity of 94.74% [95% confidence interval (CI), 73.97%–99.87%] and 100.00% (95% CI, 99.15%–100.00%) at 0.125% VAF and/or ≥5 alt reads, assuming 50 ng assay input and optimal sequencing. For follow up samples, previously discovered variants were reported at ≥2 reads, ≥0.05%.
ctDNA dynamics analysis
Additional to the QC requirements, for the predictive on-treatment ctDNA dynamics analysis, a prespecified criteria had to be met. A minimum of 14 days of treatment in the first cycle was required. Variants with a VAF <0.5% in C1D1, set to allow an above limit of detection to allow a dynamic range for on-treatment ctDNA analysis, were excluded. A ctDNA ratio (CDR) for C1D1–C1D15 was calculated per patient as the weighted mean (mean of each individual C1D15 VAF divided by the corresponding individual C1D1 VAF for each SNV or indel matching the definition above) for potentially clonal mutations at C1D1. Clonal mutations were defined as mutations with VAF > 0.5 of the maximum VAF for a patient. Alternative methodologies for calculating an optimal cutoff were also explored in this work. Briefly, a CDR based on the log-fold change [ln(C1D15+0.1)−ln(C1D1+0.1)] as a way to take into account the relative differences and a second alternative CDR using only the mutation with the maximum allele frequency on C1D1 for the ratio.
Statistics
For survival analysis, PFS was defined as the time from randomization to the first documented progressive disease (PD) based on the investigators' assessments, using RECIST v1.1, or death from any cause, whichever occurs first. Overall survival (OS) analysis was exploratory in nature, given the noncontrolled treatments upon progression on PEARL. OS was defined as the time from the date of randomization to the date of death from any cause.
Cox regression models were used in all PFS and OS analysis conducted in this study, both univariate and multivariate adjusting for known clinicopathologic variables known to be relevant in the metastatic setting (site of disease, sites of metastasis, prior chemotherapy for MBC, prior sensitivity to HT, ECOG). A P value <0.05 was considered for statistical significance.
For response analysis, objective response (OR) was defined as a complete response (CR) or partial response (PR) out of the patients who had measurable disease and Clinical Benefit Rate (CBR) as the sum of OR and stable disease (SD) according to RECIST v1.1, recorded from randomization until disease progression or death due to any cause. Tumor response was assessed on the basis of the investigator's assessment according to RECIST v1.1, and this assessment was performed at baseline and every 8 weeks (±7days) using the same method of measurement. The best response across treatment was recorded.
The optimal cutoff was selected using the minimum P value method. The procedure selects the cutoff that minimizes the significance level of the log-rank test with comparison of the two groups defined by the cutoff. The selection of the candidate cutoffs was studied only in the central range of the variable, between the 20 per cent and the 80 per cent quantile of the distribution of the continuous variable. The P value of the log-rank test using a maximally selected cutoff was adjusted for the method proposed by Lausen and Schumacher (17), with an additional permutation test assessing statistical significance of the model based in the statistic of the log-rank test (17). A Cox regression model was calculated with the binary variable obtained with the optimal cut point method, and the potential overestimation of the log-HR was corrected by a shrinkage factor c as proposed by Verweij and Van Houwelingen (18). A bootstrap resampling proposed by Hollander and Schumacher was used to calculate an adequate confidence interval of the log-HR (19). The statistical significance of the log-HR was valuated with a permutation test, repeating the whole cutpoint selection process with randomly permuted survival times and censoring (17). R v4.1.1 was used as statistical software.
Data sharing statement
The complete set of results used in the analysis, for all patients involved, is available on Supplementary Table S2 and Supplementary Table S3.
The clinical database of this study is available upon request from the corresponding author.
Results
Demographic analysis
A total of 201 patients with baseline sample for prognostic analysis were included, 107 (53%) treated in the palbociclib plus fulvestrant arm and 94 (47%) in the capecitabine arm. Our study population did not differ in baseline characteristics with the overall PEARL population (Supplementary Table S4). Analysis of classical demographic characteristics showed no significant differences between treatment, excepting more patients with multiple sites of metastasis in the capecitabine arm (76.6% vs. 59.81%; P = 0.02), but without differences in visceral versus non-visceral disease between arms (Supplementary Table S5). Similarly, PFS in this population was not significantly different between treatment arms [median PFS (mPFS) 7.82 vs. 9.26 months, m; HR, 0.97; 95% CI, 0.7–1.35; P = 0.86; Padj = 0.96; Supplementary Fig. S1A]. Exploratory OS also showed nonsignificant differences between treatments [median OS (mOS) 23.95 vs. 30.03 m; HR, 1.24; 95% CI, 0.74–2.06; P = 0.41; Padj = 0.45; Supplementary Fig. S1B].
Baseline prognostic analysis
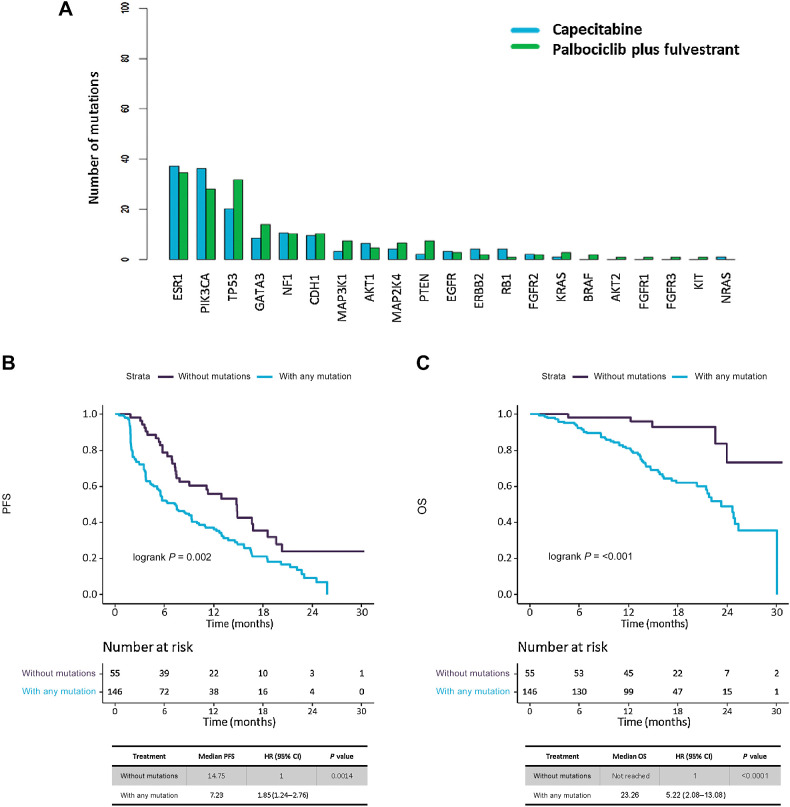
From total 201 patients, 146 (73%) had at least one mutation identified in their baseline sample (i.e., ctDNA detection), with 55 (27%) without any mutation detected (Supplementary Table S6 includes a full summary of all mutations detected at baseline for the 146 patients with ctDNA detection). Baseline genomic landscape did not differ between treatments arms (Supplementary Table S7) with ESR1, PIK3CA and TP53 being the genes most commonly mutated in both arms (Fig. 2A). PFS was significantly worse in patients with ctDNA detection versus those with no detection (mPFS 7.23 m vs. 14.75m; HR, 1.85; 95% CI, 1.24–2.76; P < 0.01; Fig. 2B). OS was also significantly worse in patients with ctDNA detection versus no detection (mOS 23.26 m vs. not reached; HR 5.22; 95% CI, 2.08–13.08; P < 0.01; Fig. 2C).
Figure 2.
Mutation distribution across study population, and analysis of ctDNA detection effect in survival. A, Bar plot showing distribution of mutations in study population, by mutated genes and treatment arms. B and C, Kaplan–Meier survival curves, including risk table (time in months, m), to measure presence of any mutation (ctDNA detection) effect in survival, among patients with and without any mutation detected. B, PFS, and (C) OS.
Baseline PFS prognostic analysis. TP53, ESR1 and PIK3CA
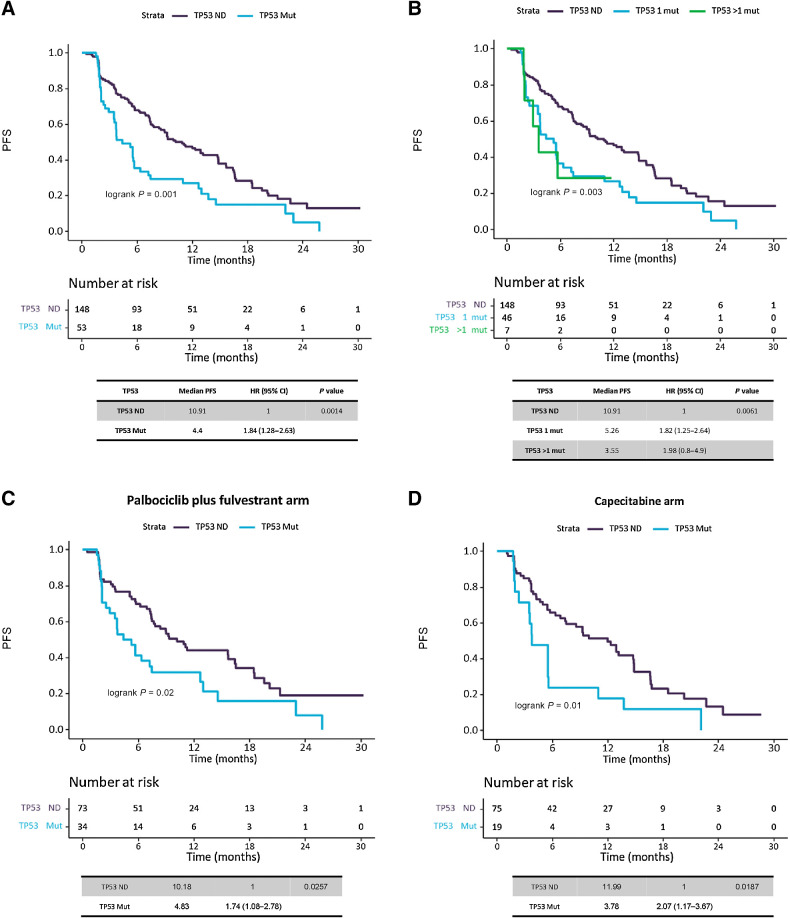
Overall, patients with a detectable TP53 mutation had worse PFS than those with TP53 mutation not detected (TP53 ND) with mPFS 4.4 m versus 10.9 m (HR, 1.84; 95% CI, 1.28–2.63; P < 0.01; Padj < 0.01; Fig. 3A). In addition, patients harboring more than one TP53 mutation had worse PFS (mPFS 3.55 m) and showed an increased HR (1.98; 95% CI, 0.8–4.9) than those with only one mutation (mPFS 5.26 m; HR, 1.82; 95% CI, 1.25–2.64), when compared with TP53 ND (mPFS 10.91 m; P < 0.01; Padj = 0.02; Fig. 3B). In the palbociclib plus fulvestrant arm, PFS was worse for patients with TP53 mutations detected vs. not detected (mPFS 4.83 vs. 10.18m; HR, 1.74; 95% CI, 1.08–2.78; P = 0.03; Padj = 0.06; Fig. 3C). In the capecitabine arm, PFS was also worse for patients with TP53 mutations detected vs. not detected (mPFS 3.78 vs. 11.99 m; HR, 2.07; 95% CI, 1.17–3.67; P = 0.02; P < 0.05; Fig. 3D). Adverse PFS conferred by multiple versus one versus TP53 ND was also apparent when analyzing separately patients on palbociclib and fulvestrant or capecitabine but with limited numbers (Supplementary Fig. S2A and S2B). There was no interaction between TP53 mutations and treatment randomization (P = 0.69; Padj = 0.63).
Figure 3.
Analysis of TP53 mutation effect in PFS. Kaplan–Meier survival curves, including risk table (time in months, m), to measure the effect of TP53 mutations in PFS. A, Survival analysis on TP53 gene mutations, regardless treatment, by presence/absence of mutations. B, Survival analysis on TP53 gene mutations, regardless treatment, by number of mutations (0 mutations, 1 mutation or more than 1 mutations). C, Survival analysis on TP53 gene mutations, in palbociclib plus fulvestrant treated patients, by presence/absence of mutations. D, Survival analysis on TP53 gene mutations, in capecitabine treated patients, by presence/absence of mutations.
Given that patients with ctDNA detection had been previously found to have significantly worse PFS compared with those without any mutation detected, we then repeated the analysis considering ctDNA detection as a surrogate of higher tumor burden and more aggressive biology and potential confounding variable; therefore, we repeated analysis removing patients without ctDNA detection. Analysis within the population with ctDNA detection also showed patients with TP53 mutation had worse PFS than those with TP53 ND with mPFS 4.4 m vs. 9.26 m (HR, 1.48; 95% CI, 1–2.17; P = 0.05; Padj = 0.10; Supplementary Fig. S3A).
No significant differences were found in PFS by ESR1 or PIK3CA mutations in analysis for both complete population and restricted to ctDNA detection. A summary of baseline ESR1 and PIK3CA mutations can be found on Supplementary Table S8. A summary of results for univariate and multivariate PFS analyses can be found on Supplementary Table S9.
Baseline OS prognostic analysis. TP53
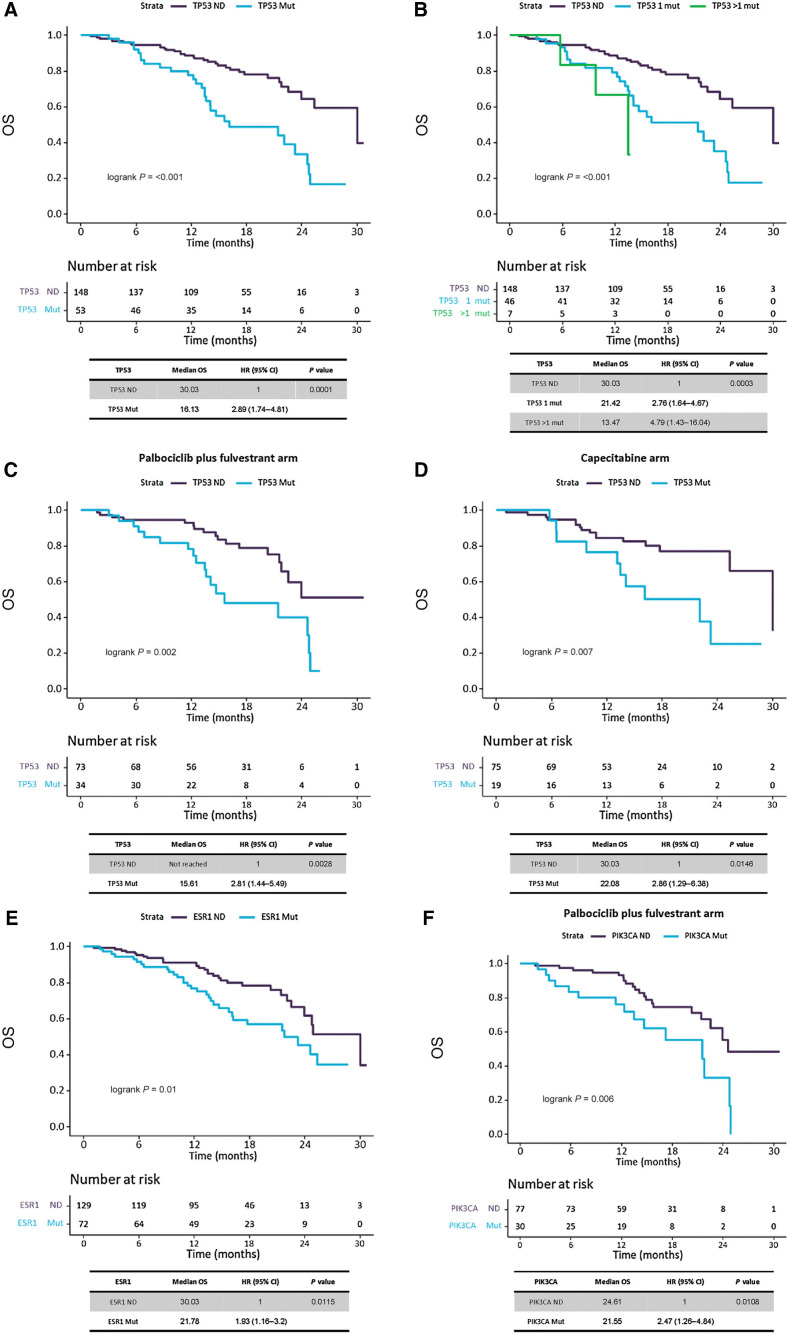
Patients with a detectable TP53 mutation had worse OS than those with TP53 ND with mOS 16.13 m versus 30.03 m (HR, 2.89; 95% CI, 1.74–4.81; P = 0.0001; Padj = 0.0004; Fig. 4A). Patients harboring more than one TP53 mutation had worse OS (mOS 13.47m) and showed an increased HR (4.79; 95% CI, 1.43–16.04) than those with only one TP53 mutation (mOS 21.42m; HR, 2.76; 95% CI, 1.64–4.67) when compared with TP53 ND (mOS 30.03m; P < 0.001; Padj = 0.001; Fig. 4B). The adverse OS risk conferred by TP53 mutation was maintained in patients treated with palbociclib plus fulvestrant (15.61 m vs. not reached; HR, 2.81; 95% CI, 1.44–5.49; P < 0.01; Padj = 0.04; Fig. 4C) and patients treated with capecitabine (22.08 m vs. 30.03; HR, 2.86; 95% CI, 1.29–6.38; P = 0.01; Padj = 0.04; Fig. 4D). Interaction test for OS between treatment and TP53 mutations was not significant (P = 0.96; Padj = 0.87).
Figure 4.
Analysis of TP53, ESR1 and PIK3CA mutation effect in OS. Kaplan–Meier survival curves, including risk table (time in months, m), to measure the effect of TP53 mutations in OS. A, Survival analysis on TP53 gene mutations, regardless treatment, by presence/absence of mutations. B, Survival analysis on TP53 gene mutations, regardless treatment, by number of mutations (0 mutations, 1 mutation or more than 1 mutations). C, Survival analysis on TP53 gene mutations, in palbociclib plus fulvestrant treated patients, by presence/absence of mutations. D, Survival analysis on TP53 gene mutations, in capecitabine treated patients, by presence/absence of mutations. E, Survival analysis on ESR1 gene mutations, regardless treatment, by presence/absence of mutations. F, Survival analysis on PIK3CA gene mutations, in palbociclib plus fulvestrant treated patients, by presence/absence of mutations.
Acknowledging the impact of ctDNA detection also in OS, we repeated our OS analysis in the population with ctDNA detection. This analysis confirmed patients with TP53 mutation had worse OS than those with TP53 ND (mOS 16.13 m vs. 30.03m; HR, 2.06; 95% CI, 1.21–3.5; P < 0.01; Padj = 0.02; Supplementary Fig. S3B), also confirming worse OS and increased HR for patients harboring more than one TP53 mutations (mOS 13.47m; HR, 3.31; 95% CI, 0.98–11.19) than those with only one mutation (mOS 21.42m; HR, 1.97; 95% CI, 1.14–3.4), when compared with TP53 ND (mOS 30.03m; P = 0.02; Padj = 0.03; Supplementary Fig. S3C).
Baseline OS prognostic analysis. ESR1 and PIK3CA
Patients with a detectable ESR1 mutation were also found to have worse OS than those with ESR1 ND with mOS 21.78 m versus 30.03 m (HR, 1.93; 95% CI, 1.16–3.2; P = 0.01; Padj = 0.04; Fig. 4E). This observation was maintained when stratifying by number of mutations, as patients with more than one ESR1 mutation showed worse OS with an increased HR (mOS 21.55 m; HR, 2.45; 95% CI, 1.27–4.72) than those with only one mutation (mOS 24.61 m; HR, 1.67; 95% CI, 0.92–3.01), when compared with ESR1 ND (mOS 30.03 m; P = 0.02, Padj = 0.07). The magnitude of OS differences was less evident when analysis was repeated by treatment allocation, with palbociclib plus fulvestrant (21.55 m vs. 24.77m; P = 0.09, Padj = 0.11) or capecitabine (23.26 m vs. 30.03 m; P = 0.04, Padj = 0.13). Interaction test for OS between treatment and ESR1 was not significant (P = 0.71; Padj = 0.77).
Analysis within population with ctDNA detection did not show differences in OS for presence of ESR1 mutation (P = 0.47; Padj = 0.99; Supplementary Fig. S4A).
Patients with a detectable PIK3CA mutation were found to have similar OS than those with PIK3CA ND in the overall population (P = 0.14). Interestingly, and unlike TP53 and ESR1, differences in OS appeared to be restricted to patients on palbociclib plus fulvestrant with mOS 21.55 m vs. 24.61 m (HR, 2.47; 95% CI, 1.26–4.84; P = 0.01; Padj = 0.02; Fig. 4F), maintained when exploring number of mutations detected [PIK3CA > 1 mutation detected (mOS 12.32 m; HR, 7.16; 95% CI, 2.78–18.47) and PIK3CA 1 mutation detected (mOS 21.78; HR, 1.74; 95% CI, 0.79–3.83)] and compared with PIK3CA ND (mOS 24.61 m; P < 0.01; Padj = 0.03). Interaction test for OS between treatment and PIK3CA was significant in the univariate (P = 0.04) and marginally significant in the multivariate analysis (P = 0.05).
Analysis within the population with ctDNA detection confirmed no overall influence in OS for PIK3CA mutations (P = 0.71, Padj = 0.75) but confirmed opposite OS trend for PIK3CA mutations when stratifying by treatment. In capecitabine treatment arm, patients with PIK3CA mutations showed higher OS than those in palbociclib and fulvestrant, resulting in a significant interaction test for OS in this selected population between treatment and PIK3CA (P < 0.01; Padj < 0.01; Supplementary Fig. S4B–S4D).
A summary of results for univariate and multivariate OS analyses in ESR1 and PIK3CA mutations can be found in Supplementary Table S10.
Predictive ctDNA dynamic analysis
A total of 120 patients met prespecified criteria for assessing ctDNA dynamics, C1D1-C1D15 CDR analysis, 64 (53%) in the palbociclib plus fulvestrant arm and 56 (47%) in the capecitabine arm. From the overall baseline population, 81 (40%) patients were not included in the CDR analysis because of baseline ctDNA not detected (n = 52; 64%), baseline mutations only <0.5% VAF (n = 14; 17%), not having a paired C1D15 sample (n = 14; 17%) and having a C1D15 sample failing sequencing (n = 1; 1%; Fig. 1). Patients for CDR analysis had significantly more ECOG 1 than non-CDR patients, without other clinicopathologic differences (Supplementary Table S11). Significant differences in survival were found between CDR and non-CDR population (mPFS 6.21 m vs. 14.75 m, respectively) with increased HR in CDR population (HR, 1.78; 95% CI, 1.25–2.52; P < 0.001; Supplementary Fig. S1C), consistent with the fact that 81% of the non-CDR had no baseline ctDNA detection or very low frequency mutations (<0.5% VAF). There were no differences in genomic landscape between CDR and non-CDR populations (Supplementary Table S12).
CDR and PFS
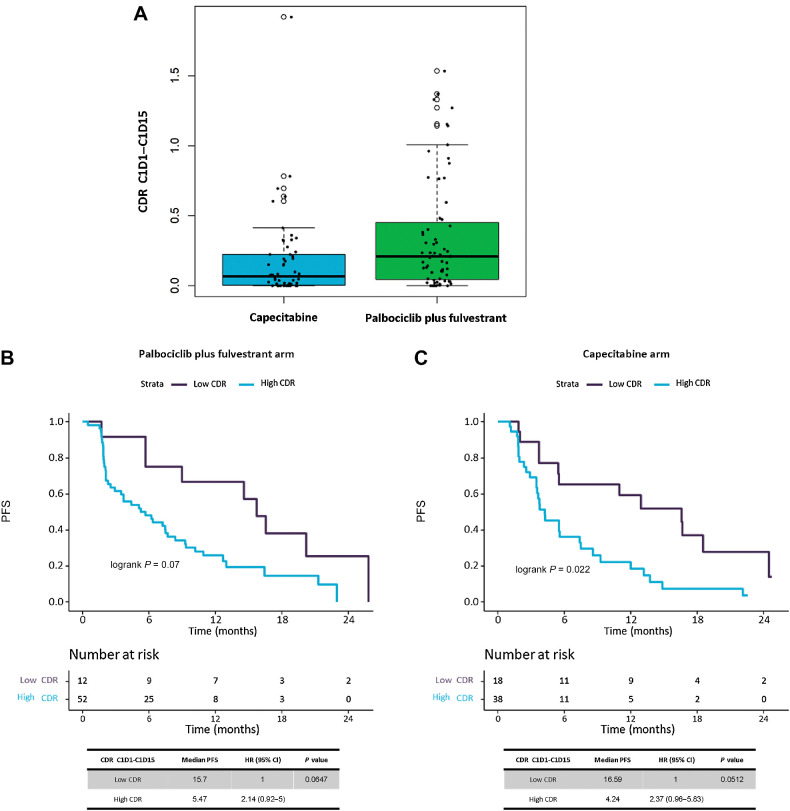
Median CDR between C1D15–C1D1 calculated with prespecified methodology was significantly different between treatment arms, with patients treated with capecitabine having a lower CDR (i.e., more profound ctDNA suppression) than those on palbociclib plus fulvestrant (median CDR 0.07 vs. 0.21; P < 0.01; Fig. 5A). Furthermore, a greater proportion of patients treated with capecitabine had a CDR = 0 (i.e., complete ctDNA suppression under limit of detection) compared with those on palbociclib plus fulvestrant (23.21% vs. 7.81%, respectively; P = 0.02).
Figure 5.
Longitudinal predictive ctDNA analysis by use of median CDR methodology, calculated at C1D15 timepoint. A, Boxplot graph showing distribution of CDR C1D15 values in patients by treatment arms, palbociclib plus fulvestrant vs. capecitabine. B and C, Kaplan–Meier survival curves, including risk table (time in months, m), to measure the effect of CDR (Low vs. High) in PFS by treatment arm, (B) in palbociclib plus fulvestrant treated patients and (C) in capecitabine treated patients. Median CDR between C1D15–C1D1, calculated with prespecified methodology, offered a CDR value optimal cutoff of 0.0247 for palbociclib plus fulvestrant arm, while it was of 0.0127 for the capecitabine treatment arm. Those optimal cut-off values were used to create Low versus High CDR groups, in both treatment arms.
We then tested how our proposed optimal methodology could be used to predict PFS in each treatment arm. In the palbociclib plus fulvestrant arm optimal cutoff was defined at CDR = 0.0247, with HR 2.14 (95% CI, 0.92–5), P = 0.06 (Fig. 5B). In the capecitabine arm optimal cutoff was 0.0127, with HR 2.37 (95% CI, 0.96–5.83), P = 0.05 (Fig. 5C).
Analysis of potential detrimental effect of TP53 mutations in patients with ctDNA suppression, although limited in number, showed no significant differences in survival in both treatment arms with overall improved survival in patients with TP53 mutations and ctDNA suppression, suggesting patients achieving early ctDNA drop surpassed the detrimental effect of baseline TP53 mutations (palbociclib plus fulvestrant: mPFS 14.55 vs. 16.53 m; HR, 1; 95% CI, 02–5; P = 0.99 and capecitabine: mPFS 8.25 vs. 16.66 m; HR, 2.71; 95% CI, 0.52–14.06; P = 0.27).
A weighted mean for clonal mutations at C1D1 was the best methodology for predictions in both treatment arms. Although inferior, alternative methodologies explored for CDR calculations and predictions showed overall relatively similar results (Supplementary Table S13).
CDR and response
Response data were available in 118 patients of the 120 total patients included into the longitudinal predictive ctDNA analysis. OR defined as a CR or PR and CBR as the sum of OR and SD according to RECIST v1.1, were evaluated according to CDR values for all patients included in the study.
OR was 18% (all responses recorded as PR), and CBR was 73.7%. Patients under PD represented 26.3%.
We found a significant association of greater CDR suppression overall in patients with CBR (N = 87) versus those progressing (N = 31) with CDR 0.1 versus 0.2, respectively (P = 0.03), but differences were not maintained when separated per treatment (P = 0.12 for palbociclib plus fulvestrant and P = 0.29 for capecitabine), however with limited sample size (Supplementary Fig. S5).
Discussion
We show that a significant proportion of patients with HR+/HER2− MBC have a detectable TP53 mutation in plasma and this confers them a significantly worse outcome compared with those patients where the mutation was not detected. Patients with TP53 mutations had very poor survival in both endocrine-based or chemotherapy treatment arms, extending results reported in patients from PALOMA-3 trial treated with endocrine-based therapies (9), and suggest aggressive behaviour irrespectively of treatment, highlighting the need of designing clinical strategies to improve outcomes in this population (20). We believe TP53 mutations should be incorporated in future prognostic and predictive models for HR+/HER2− MBC given this deleterious effect could be sufficiently strong to prevent applicability of models not incorporating this information. In addition, in our exploratory OS analysis, TP53 mutations also conferred patients a worse OS. Although subsequent treatments were not controlled upon progression on PEARL, we now have evidence that both PFS and OS did not differ between treatment arms (21) suggesting consistency of our OS impact finding. Interestingly, we found that multiple TP53 mutations confer even worse outcomes, an observation that would need further validation. We acknowledge a potential limitation for these findings is the fact that we did not perform germline correction for our sequencing and that some TP53 mutations identified could be resulting from clonal hematopoiesis (22, 23). Given the high incidence of TP53 mutations in breast cancer, clonal hematopoiesis may have played some contamination effect in our conclusions. However, we sought to take some pre-planned actions to minimize this effect. Apart from usual QC, known population-level germline variants were eliminated as per variant population frequency. We further performed analysis and filtering of the remaining data using custom R scripts of potential germline mutations based on VAF and only oncogenic/likely oncogenic variants were included in our analysis. TP53 mutations are present in 5% to 10% of clonal hematopoeiesis of indeterminate potential (CHIP) (24, 25) and maximum estimates of the incidence of detectable CHIP with ctDNA in patients with MBC are 25% patients (26). Therefore, only 1.25% to 2.5% of patients in our study would be expected to have potential TP53 CHIP mutations and most of them would have been filtered by our analysis methods. We therefore believe CHIP would have had a minimal effect in our conclusions. We acknowledge germline white blood cell DNA sequencing would have helped optimize CHIP substraction, but unfortunately this was not possible in our study. Other frequent mutations present or acquired in this population, like ESR1 or PIK3CA mutations, were particularly relevant in the exploratory OS analyses. ESR1 mutations were found to confer worse survival matching previous observations (16, 27, 28). In addition, a detrimental effect of PIK3CA mutations was also found for OS on those patients treated with palbociclib and fulvestrant. In particular, this reinforces the growing evidence that PIK3CA mutations detected in MBC seem to confer worse outcomes as opposed to detection in early settings, where it has been traditionally associated with better outcomes (29). Proportion of patients with ESR1 mutation was slightly higher in or study compared with previous reports in studies conducted in tissue samples, likely reflecting the effectiveness of ctDNA assays to pick up this particular mutation, often polyclonal and present at the sub-clonal level.
For our early on-treatment predictions, we applied a previously reported (15) prespecified methodology for our CDR calculations, consisting on a weighted mean for VAFs in genes considered potentially clonal in the pretreatment sample. We show that this methodology can select patients deriving an optimal benefit for both treatment arms of our study, adding evidence to previous publications identifying early ctDNA dynamics as a potentially useful tool for efficacy predictions (11–14).
We acknowledge our CDR results have to be considered exploratory and further validation is required. Our panel may seem limited to broadly explore ctDNA dynamics, but our methodology seeks to measure early dynamics in relevant mutations with enough representation in patients’ blood circulation to be reliably measured for change, an important issue not fully addressed in other similar reports. We also acknowledge our approach is potentially less sensitive than a tumor-informed approach, but given we focus on change, we think the critical point is having a reliable quantification of mutation abundance (i.e., VAF in our study), and we sought to accomplish this by focusing on relevant genes and performing a deep error-corrected sequencing. We believe our CDR results are limited, yet we attempted to provide rigorous data. An optimal cutoff for treatment efficacy predictions is in nature context-dependent and will vary between clinical setting, treatment, and algorithm used to measure the ctDNA change. However, it is interesting to notice that optimal cutoffs found in PEARL with our methodology are in line with those reported in PALOMA-3 (11) and previous reports with subsets of patients treated with targeted therapies (15), suggesting an important early ctDNA suppression often in the range of 0 to 0.1 CDR is required to select those patients deriving the greatest benefit. We notice that this may not apply to patients treated with immunotherapies, where less ctDNA suppression may be sufficient to select good responders (30). We hypothesize that an important degree of cytostatic or cytotoxic effect must be achieved with both endocrine-based or chemotherapy treatments to predict long term benefit, whereas for patients on immunotherapy microenvironment plays an important role in controlling proliferation and less cytostatic or cytotoxic effect reflected in higher CDR might be sufficient to select optimal responders.
ctDNA suppression on day 15 was significantly more pronounced in patients treated with capecitabine versus those on palbociclib and fulvestrant, suggesting chemotherapy cytotoxicity might result in faster suppression.
Interestingly, patients not having any detectable baseline mutation had significantly better survival compared with those with evaluable mutations. This is expected and adds evidence towards not having a detectable mutation probably reflecting low tumor burden or proliferation and therefore better prognosis independent of the treatment received.
In summary, our work shows impaired PFS irrespective of endocrine or chemotherapy-based treatments for patients with HR+/HER2− MBC harboring plasma TP53 mutations. Our findings also suggest presence of plasma ESR1 mutations could be detrimental in the long term for these patients and adds additional evidence that early ctDNA suppression may be clinically helpful for long-term treatment efficacy predictions, suggesting an optimal methodology that helps selecting patients deriving the most benefit. Further validation to fully demonstrate clinical utility of ctDNA dynamics is warranted.
Supplementary Material
Supplementary Figure 1. Survival analysis in baseline population (C1D1).
Supplementary Figure 2. PFS analysis in baseline population (C1D1) on TP53 mutations, by treatment arms.
Supplementary Figure 3. Survival analysis in baseline population (C1D1) on TP53 mutations, within the population with any detectable mutation (n=146).
Supplementary Figure 4. OS analysis in baseline population (C1D1) on ESR1 and PIK3CA mutations, within the population with any detectable mutation (N=146).
Supplementary Figure 5. CDR and clinical benefit rate within the longitudinal predictive ctDNA analysis.
Supplementary Table 1. List of the 21 genes included in the breast cancer panel used for sequencing, and the targeted coding regions in each case.
Supplementary Table 2. Complete table results for all patients (n=201) involved in baseline (C1D1) analysis.
Supplementary Table 3. Complete table results for all patients (n=120) involved in C1D15 analysis.
Supplementary Table 4. Baseline characteristics comparison between our selected study population and the overall PEARL population.
Supplementary Table 5. Baseline characteristics comparison between treatment arms in the selected study population
Supplementary Table 6. Summary of all baseline mutations identified for the 146 patients with ctDNA detection.
Supplementary Table 7. Baseline genomic landscape of mutations distribution in population, by number of mutations or presence/absence, across treatment arms.
Supplementary Table 8. Summary of baseline ESR1 and PIK3CA mutations, with number of affected patients and mean VAF.
Supplementary Table 9. Summary of PFS results for ESR1 and PIK3CA mutations. Cox regression univariate and multivariate analysis.
Supplementary Table 10. Summary of OS results for ESR1 and PIK3CA mutations. Cox regression univariate and multivariate analysis.
Supplementary Table 11. Baseline characteristics comparison between population with CDR calculation (n=120) vs non-CDR population (n=81).
Supplementary Table 12. Baseline genomic landscape of mutations distribution for the CDR population (n=120), by number of mutations or presence/absence, across treatment arms.
Supplementary Table 13. Alternative methodologies used for the calculation of CDR and PFS predictions.
Acknowledgments
This work was supported by two funding companies: Pfizer Inc. (that provided palbociclib and exemestane through a collaboration agreement) and AstraZeneca (that provided fulvestrant). The trial sponsor is GEICAM Spanish Breast Cancer Group. The study was as well supported with donation from the patient association AMUMA (Asociación de Cáncer de Mama y Ginecológico de Castilla-La Mancha).
We thank all the patients included in this study and their families, as well as all the participating investigators and the support staff at each study site and at both the GEICAM and CECOG headquarters.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
J. Pascual reports personal fees and nonfinancial support from AstraZeneca; personal fees from Novartis; and nonfinancial support from Pfizer outside the submitted work. M. Gil-Gil reports personal fees from Pfizer, Novartis; and other support from Lilly outside the submitted work. C. Zielinski reports grants from AstraZeneca, Pfizer, Servier, Novartis, Eli Lilly, Roche, BMS, MSD, Takeda, Daiichi Sankyo, Celgene, Halozyme, Amgen, and Boehringer Ingelheim during the conduct of the study; in addition, C. Zielinski has a patent for HerVaxx issued and licensed to Imugene. E.M. Ciruelos Gil reports personal fees from Lilly, Roche, AstraZeneca, Daiichi Sankyo, Seagen, Gilead, Novartis, Pfizer, and MSD during the conduct of the study. M. Muñoz-Mateu reports personal fees from Roche, Gilead, Segean, AstraZeneca, Gilead, and Lilly outside the submitted work. B. Bermejo reports personal fees from Pfizer; grants from Novartis, AstraZeneca, Daiichi Sankyo, and MSD outside the submitted work. M. Margeli Vila reports grants from Pfizer; other support from Novartis, Gylead, PiereFabre, Lilly, and Daiichi Sankyo outside the submitted work. A. Antón reports personal fees from Pfizer during the conduct of the study and personal fees from Gilead, Eli Lilly, Daiichi Sankyo, Seagen, and AstraZeneca outside the submitted work. Y. Liu reports other support from Pfizer outside the submitted work. M. Hubank reports other support from Guardant Health; personal fees from Roche, Qiagen, Novartis, GSK, Bayer, Incyte, Jansenn, and AstraZeneca outside the submitted work. N.C. Turner reports has received advisory board honoraria from AstraZeneca, Lilly, Pfizer, Roche/Genentech, Novartis, GlaxoSmithKline, Repare Therapeutics, Relay Therapeutics, Zentalis, Gilead, Inivata, Guardant, Exact Sciences; and research funding from AstraZeneca, Pfizer, Roche/Genentech, Merck Sharpe and Dohme, Guardant Health, Invitae, Inivata, Personalis, and Natera. M. Martín reports personal fees from Pfizer, Puma, Roche, Novartis, Daiichi Sankyo, Seagen, Lilly, and AstraZeneca during the conduct of the study; and personal fees from SANOFI outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
J. Pascual: Conceptualization, data curation, formal analysis, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M. Gil-Gil: Resources, investigation, writing–review and editing. P. Proszek: Data curation, formal analysis, methodology, writing–review and editing. C. Zielinski: Investigation, writing–review and editing. A. Reay: Data curation, formal analysis, methodology, writing–review and editing. M. Ruiz-Borrego: Resources, investigation, writing–review and editing. R. Cutts: Data curation, formal analysis, methodology, writing–review and editing. E.M. Ciruelos Gil: Resources, investigation, writing–review and editing. A. Feber: Data curation, formal analysis, methodology, writing–review and editing. M. Muñoz-Mateu: Resources, investigation, writing–review and editing. C. Swift: Data curation, formal analysis, methodology, writing–review and editing. B. Bermejo: Resources, investigation, writing–review and editing. J. Herranz: Data curation, formal analysis, methodology, writing–original draft, writing–review and editing. M. Margeli Vila: Resources, investigation, writing–review and editing. A. Antón: Resources, investigation, writing–review and editing. Z. Kahan: Resources, investigation, writing–review and editing. T. Csöszi: Resources, investigation, writing–review and editing. Y. Liu: Funding acquisition, writing–review and editing. D. Fernandez Garcia: Data curation, formal analysis, methodology, writing–original draft, project administration, writing–review and editing. I. Garcia-Murillas: Data curation, formal analysis, methodology, writing–review and editing. M. Hubank: Conceptualization, methodology, writing–review and editing. N.C. Turner: Conceptualization, formal analysis, funding acquisition, methodology, writing–original draft, project administration, writing–review and editing. M. Martín: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Finn RS, Martin M, Rugo HS, Jones S, Im S-A, Gelmon K, et al. Palbociclib and letrozole in advanced breast cancer. N Engl J Med 2016;375:1925–36. [DOI] [PubMed] [Google Scholar]
- 2. Hortobagyi GN, Stemmer SM, Burris HA, Yap Y-S, Sonke GS, Paluch-Shimon S, et al. Ribociclib as first-line therapy for HR-positive, advanced breast cancer. N Engl J Med 2016;375:1738–48. [DOI] [PubMed] [Google Scholar]
- 3. Goetz MP, Toi M, Campone M, Sohn J, Paluch-Shimon S, Huober J, et al. MONARCH 3: abemaciclib as initial therapy for advanced breast cancer. J Clin Oncol 2017;35:3638–46. [DOI] [PubMed] [Google Scholar]
- 4. Turner NC, Ro J, André F, Loi S, Verma S, Iwata H, et al. Palbociclib in hormone receptor–positive advanced breast cancer. N Engl J Med 2015;373:209–19. [DOI] [PubMed] [Google Scholar]
- 5. Slamon DJ, Neven P, Chia S, Fasching PA, De Laurentiis M, Im S-A, et al. Phase III randomized study of ribociclib and fulvestrant in hormone receptor–positive, human epidermal growth factor receptor 2–negative advanced breast cancer: MONALEESA-3. J Clin Oncol 2018;36:2465–72. [DOI] [PubMed] [Google Scholar]
- 6. Sledge GW, Toi M, Neven P, Sohn J, Inoue K, Pivot X, et al. MONARCH 2: abemaciclib in combination with fulvestrant in women with HR+/HER2−advanced breast cancer who had progressed while receiving endocrine therapy. J Clin Oncol 2017;35:2875–84. [Google Scholar]
- 7. Martin M, Zielinski C, Ruiz-Borrego M, Carrasco E, Turner N, Ciruelos EM, et al. Palbociclib in combination with endocrine therapy versus capecitabine in hormonal receptor-positive, human epidermal growth factor 2-negative, aromatase inhibitor–resistant metastatic breast cancer: a phase III randomized controlled trial—PEARL. Ann Oncol 2021;32:488–99. [DOI] [PubMed] [Google Scholar]
- 8. Rostami A, Lambie M, Yu CW, Stambolic V, Waldron JN, Bratman SV. Senescence, necrosis, and apoptosis govern circulating cell-free DNA release kinetics. Cell Rep 2020;31:107830. [DOI] [PubMed] [Google Scholar]
- 9. O'Leary B, Cutts RJ, Huang X, Hrebien S, Liu Y, André F, et al. Circulating tumor DNA markers for early progression on fulvestrant with or without palbociclib in ER+ advanced breast cancer. J Natl Cancer Inst 2021;113:309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tolaney SM, Toi M, Neven P, Sohn J, Grischke E-M, Llombart-Cussac A, et al. Clinical significance of PIK3CA and ESR1 mutations in circulating tumor DNA: analysis from the MONARCH 2 study of abemaciclib plus fulvestrant. Clin Cancer Res 2022;28:1500–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun 2018;9:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darrigues L, Pierga J-Y, Bernard-Tessier A, Bièche I, Silveira AB, Michel M, et al. Circulating tumor DNA as a dynamic biomarker of response to palbociclib and fulvestrant in metastatic breast cancer patients. Breast Cancer Res 2021;23:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martínez-Sáez O, Pascual T, Brasó-Maristany F, Chic N, González-Farré B, Sanfeliu E, et al. Circulating tumor DNA dynamics in advanced breast cancer treated with CDK4/6 inhibition and endocrine therapy. NPJ breast cancer 2021;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pascual J, Lim JSJ, Macpherson IR, Armstrong AC, Ring A, Okines AFC, et al. Triplet therapy with palbociclib, taselisib, and fulvestrant in PIK3CA-mutant breast cancer and doublet palbociclib and taselisib in pathway-mutant solid cancers. Cancer Discov 2021;11:92–107. [DOI] [PubMed] [Google Scholar]
- 15. Pascual J, Cutts RJ, Kingston B, Hrebien S, Kilburn LS, Kernaghan S, et al. Abstract PS5–02: Assessment of early ctDNA dynamics to predict efficacy of targeted therapies in metastatic breast cancer: Results from plasmaMATCH trial. Cancer Res 2021;81:PS5–02–PS05–02. [Google Scholar]
- 16. Fribbens C, O'Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, et al. Plasma ESR1 mutations and the treatment of estrogen receptor–positive advanced breast cancer. J Clin Oncol 2016;34:2961–8. [DOI] [PubMed] [Google Scholar]
- 17. Lausen B, Schumacher M. Maximally selected rank statistics. Biometrics 1992;48:73–85. [Google Scholar]
- 18. Verweij PJ, Van Houwelingen HC. Cross-validation in survival analysis. Stat Med 1993;12:2305–14. [DOI] [PubMed] [Google Scholar]
- 19. Holländer N, Sauerbrei W, Schumacher M. Confidence intervals for the effect of a prognostic factor after selection of an 'optimal' cutpoint. Stat Med 2004;23:1701–13. [DOI] [PubMed] [Google Scholar]
- 20. Hassin O, Oren M. Drugging p53 in cancer: one protein, many targets. Nat Rev Drug Discov 2023;22:127–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martín M, Zielinski C, Ruiz-Borrego M, Carrasco E, Ciruelos EM, Muñoz M, et al. Overall survival with palbociclib plus endocrine therapy versus capecitabine in postmenopausal patients with hormone receptor–positive, HER2-negative metastatic breast cancer in the PEARL study. Eur J Cancer 2022;168:12–24. [DOI] [PubMed] [Google Scholar]
- 22. Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niroula A, Sekar A, Murakami MA, Trinder M, Agrawal M, Wong WJ, et al. Distinction of lymphoid and myeloid clonal hematopoiesis. Nat Med 2021;27:1921–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kessler MD, Damask A, O'Keeffe S, Banerjee N, Li D, Watanabe K, et al. Common and rare variant associations with clonal hematopoiesis phenotypes. Nature 2022;612:301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hrebien S, Citi V, Garcia-Murillas I, Cutts R, Fenwick K, Kozarewa I, et al. Early ctDNA dynamics as a surrogate for progression-free survival in advanced breast cancer in the BEECH trial. Ann Oncol 2019;30:945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mosele F, Stefanovska B, Lusque A, Tran Dien A, Garberis I, Droin N, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol 2020;31:377–86. [DOI] [PubMed] [Google Scholar]
- 28. Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol 2016;2:1310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zardavas D, te Marvelde L, Milne RL, Fumagalli D, Fountzilas G, Kotoula V, et al. Tumor PIK3CA genotype and prognosis in early-stage breast cancer: a pooled analysis of individual patient data. J Clin Oncol 2018;36:981–90. [DOI] [PubMed] [Google Scholar]
- 30. Zhang Q, Luo J, Wu S, Si H, Gao C, Xu W, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov 2020;10:1842–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Survival analysis in baseline population (C1D1).
Supplementary Figure 2. PFS analysis in baseline population (C1D1) on TP53 mutations, by treatment arms.
Supplementary Figure 3. Survival analysis in baseline population (C1D1) on TP53 mutations, within the population with any detectable mutation (n=146).
Supplementary Figure 4. OS analysis in baseline population (C1D1) on ESR1 and PIK3CA mutations, within the population with any detectable mutation (N=146).
Supplementary Figure 5. CDR and clinical benefit rate within the longitudinal predictive ctDNA analysis.
Supplementary Table 1. List of the 21 genes included in the breast cancer panel used for sequencing, and the targeted coding regions in each case.
Supplementary Table 2. Complete table results for all patients (n=201) involved in baseline (C1D1) analysis.
Supplementary Table 3. Complete table results for all patients (n=120) involved in C1D15 analysis.
Supplementary Table 4. Baseline characteristics comparison between our selected study population and the overall PEARL population.
Supplementary Table 5. Baseline characteristics comparison between treatment arms in the selected study population
Supplementary Table 6. Summary of all baseline mutations identified for the 146 patients with ctDNA detection.
Supplementary Table 7. Baseline genomic landscape of mutations distribution in population, by number of mutations or presence/absence, across treatment arms.
Supplementary Table 8. Summary of baseline ESR1 and PIK3CA mutations, with number of affected patients and mean VAF.
Supplementary Table 9. Summary of PFS results for ESR1 and PIK3CA mutations. Cox regression univariate and multivariate analysis.
Supplementary Table 10. Summary of OS results for ESR1 and PIK3CA mutations. Cox regression univariate and multivariate analysis.
Supplementary Table 11. Baseline characteristics comparison between population with CDR calculation (n=120) vs non-CDR population (n=81).
Supplementary Table 12. Baseline genomic landscape of mutations distribution for the CDR population (n=120), by number of mutations or presence/absence, across treatment arms.
Supplementary Table 13. Alternative methodologies used for the calculation of CDR and PFS predictions.