Abstract
Purpose:
We present the results of a post hoc tumor tissue analysis from the phase 3 MILO/ENGOT-ov11 study (NCT01849874).
Patients and Methods:
Mutation/copy-number analysis was performed on tissue obtained pre-randomization. The Kaplan–Meier method was used to estimate progression-free survival (PFS). Unbiased univariate analysis, Cox regression, and binary logistic regression were used to test associations between mutation status and outcomes, including PFS and binary response by local RECIST 1.1.
Results:
MILO/ENGOT-ov11 enrolled 341 patients, ranging in age from 22 to 79, from June, 2013 to April, 2016. Patients were randomized 2:1 to binimetinib or physician's choice of chemotherapy (PCC). The most commonly altered gene was KRAS (33%). In 135 patients treated with binimetinib with response rate (RR) data, other detected MAPK pathway alterations included: NRAS (n = 11, 8.1%), BRAF V600E (n = 8, 5.9%), RAF1 (n = 2, 1.5%), and NF1 (n = 7, 5.2%). In those with and without MAPK pathway alterations, the RRs with binimetinib were 41% and 13%, respectively. PFS was significantly longer in patients with, compared with those without, MAPK pathway alterations treated with binimetinib [HR, 0.5; 95% confidence interval (CI) 0.31–0.79]. There was a nonsignificant trend toward PFS improvement in PCC-treated patients with MAPK pathway alterations compared with those without (HR, 0.82; 95% CI, 0.43–1.59).
Conclusions:
Although this hypothesis-generating analysis is limited by multiple testing, higher RRs and longer PFS were seen in patients with low-grade serous ovarian cancer (LGSOC) treated with binimetinib, and to a lesser extent in those treated with PCC, who harbored MAPK pathway alterations. Somatic tumor testing should be routinely considered in patients with LGSOC and used as a future stratification factor.
Translational Relevance.
To our knowledge, this is the largest randomized study of patients with low-grade serous ovarian cancer (LGSOC). Although the primary endpoint of this study was not met, the data generated provide valuable information regarding how molecular alterations can affect outcomes. We report a comprehensive analysis of the association between MAPK pathway alterations and patient outcomes. Higher response rates and significantly longer progression-free survival were observed in patients with LGSOC treated with binimetinib, and to a lesser extent in those treated with physician's choice of chemotherapy, who harbored MAPK pathway alterations. The results of this analysis provide reliable evidence that MAPK pathway alteration status has prognostic implications for patients with LGSOC. Somatic tumor testing should be considered for all patients with recurrent LGSOC to aid in clinical decision making regarding the relative benefit of systemic therapy and used as a stratification factor in future prospective studies of LGSOC.
Introduction
Low-grade serous ovarian cancers (LGSOC), which account for 5% to 10% of all epithelial ovarian cancers, are characterized by a protracted clinical course, p53 wild-type (WT) expression, and a high prevalence of MAPK pathway alterations; in contrast, high-grade serous ovarian cancers almost universally display aberrant p53 expression and rarely have identifiable MAPK pathway alterations (1, 2). The MAPK pathway regulates cellular proliferation through a signal transduction cascade mediated by Ras family members (KRAS, NRAS, and HRAS), resulting in activation of downstream RAF/MEK/ERK effectors, and can be inhibited by negative regulators of the pathway (3). KRAS mutations are found in 19% to 36% of LGSOC tumors, and BRAF mutations are found in 2% to 16% (4–7). Low response rates to chemotherapy, as reported in retrospective reports, as well as the unique molecular profile of LGSOC, have led to considerable interest in the use of MEK inhibitors for the treatment of patients with recurrent LGSOC (8, 9).
Findings from a phase 2 study of the oral MEK inhibitor selumetinib showed promising activity in a molecularly unselected population of patients with recurrent LGSOC, with an objective response rate (ORR) of 15.4% (10). The study showed no association between mutation status and response to selumetinib; however, this finding was limited, as only 65% of enrolled patients had sufficient DNA for genetic analysis and the study was focused specifically on detection of codon 600 mutations in BRAF and codon 12 and 13 mutations in KRAS. Subsequently, two large studies of MEK inhibitors for the treatment of recurrent LGSOC were performed, and both studies enrolled patients without molecular selection. A randomized phase 2/3 study of trametinib versus physician's choice of chemotherapy (PCC) or endocrine therapy showed a response rate of 26.2% with single-agent trametinib in women with recurrent LGSOC (GOG-0281, NCT02101788; ref. 11). The phase 3 MILO/ENGOT-ov11 study of binimetinib versus PCC for the treatment of patients with recurrent LGSOC closed prematurely after an interim analysis of the initial 303 patients (January, 2016 data cutoff date) found the HR for progression-free survival (PFS) crossed the predefined futility boundary; at the time of the interim analysis, the response rate to binimetinib was reported at 16% (12). Patients were notified of the interim analysis results and were allowed to continue binimetinib treatment until discontinuation criteria were met. An updated analysis with a January, 2019 data cutoff date using local RECIST 1.1 radiology reads showed a response rate of 24% with single-agent binimetinib (12). Trametinib and binimetinib are now category 2A and B National Comprehensive Cancer Network compendium listed, respectively, as treatment options for patients with recurrent LGSOC (13). Although these studies showed promising response rates to single-agent MEK inhibition in the general population of women with recurrent LGSOC, they also prompted significant interest in identifying biomarkers to better select patients most likely to benefit from treatment with a MEK inhibitor. Here, we present the results of the tumor tissue genetic testing performed in conjunction with the phase 3 MILO/ENGOT-ov11 study.
Patients and Methods
Original study design
The MILO/ENGOT-ov11 study enrolled women with low-grade serous cancer of the ovary or primary peritoneum. The median age of patients treated with binimetinib was 51.6 years (range, 23–79 years), and the median age of patients treated with PCC was 50.2 years (range, 22–78 years; ref. 12). In short, eligible patients had measurable recurrent or persistent disease and had received ≥1 prior platinum-based chemotherapy regimens but ≤3 chemotherapy regimens in total, with no limit to the number of lines of prior hormonal therapy. Patients had an Eastern Cooperative Oncology Group performance status of 0 or 1. Patients were excluded if they had received previous treatment with a MEK or BRAF inhibitor. Patients were randomized 2:1 to receive binimetinib or physician's choice of intravenous chemotherapy (pegylated liposomal doxorubicin, paclitaxel, or topotecan). Binimetinib was administered 45 mg orally twice daily continuously starting on day 1.
The study was approved by the institutional review board of each site. All clinical work was conducted in compliance with current Good Clinical Practices as referenced in the International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use. All patients enrolled in the study provided written, informed consent to study-directed treatment and molecular analysis of submitted tissue before their participation. The primary endpoint was to assess PFS as determined by blinded independent central review, with the local investigator assessments used as supportive analyses, which was previously reported (12). Formalin-fixed, paraffin-embedded (FFPE) tissue was collected from participants at study entry for a predefined exploratory analysis to determine alterations in cancer-associated genes based on the biomarker plan. The biomarker plan entailed that following confirmation of a low-grade serous carcinoma diagnosis, tumor tissue would be assessed for mutations and copy-number variations in cancer-associated (including MAPK pathway) genes by next-generation sequencing.
Molecular/biomarker analysis
Enrolled patients were required to submit archival tissue or undergo fresh biopsy for central pathology review before randomization. Following confirmation of diagnosis, if sufficient tumor tissue remained, a next-generation sequencing assay was used to assess a panel of 315 genes using FoundationOne (Supplementary Table S1). This post hoc analysis is based on the results of patient outcomes as of the final data cutoff date of August, 2021 and the molecular results obtained from tissue submitted for central review at the time of study entry. Unbiased univariate analysis was used to test the association between altered genes (in >5% of cohorts) and outcomes when treated with binimetinib or PCC. Examined outcomes were PFS (estimated by the Kaplan–Meier method) and binary response by RECIST 1.1 [complete response or partial response (CR/PR) vs. stable or progressive disease (SD/PD)]. Cox regression was used to assess association between mutated genes and PFS; Fisher exact tests were used to examine univariate association between mutated genes and binary response. An FDR was used to assess effect of multiple comparisons. Multivariable genomic models were constructed for outcomes with genes that had univariate P values of <0·25 and stayed independently significant at a P value of <0·05 within a Cox regression model for PFS and a binary logistic regression model for overall response. MAPK pathway alterations of interest were determined on the basis of a review of the literature (9, 11, 14, 15). Cox and binary logistic regressions were used to examine the relationship between KRAS mutation status, MAPK pathway alteration status, and outcomes. Interactions on the multiplicative and additive scales between KRAS/MAPK pathway alterations and treatment were assessed using interaction terms in full cohort PFS and OR models (16). Survival estimates stratified by KRAS mutation status and MAPK pathway alteration status were calculated with the Kaplan–Meier method. All analyses were performed in R Statistical Software (Version 4·1·3).
Data availability statement
The raw sequencing data for this study were generated at Foundation Medicine; requests for sequencing data can be sent to clinicaltrials@foundationmedicine.com. Derived data supporting the findings of this study are available from the corresponding author upon request.
Results
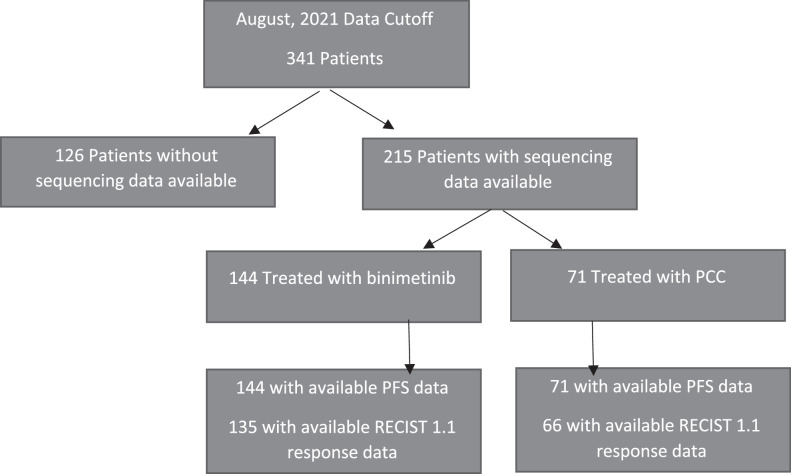
The phase 3 MILO/ENGOT-ov11 study enrolled 341 patients between June, 2013 and April, 2016. Among patients treated with binimetinib who had sequencing data available, there were 144 patients with PFS data and 135 patients with RECIST 1.1 response data. Among patients treated with PCC who had molecular analysis data available, there were 71 patients with PFS data and 66 patients with RECIST 1.1 response data (Fig. 1). Forty-seven genes among those tested in the gene panel were altered in at least 5% of patients. KRAS was the most common altered gene, occurring in 33% of patients. The frequency of KRAS mutation was similar between the two groups—46 patients (32%) treated with binimetinib and 24 patients (34%) treated with PCC (see Supplementary Table S2 for the distribution of KRAS mutations and other MAPK pathway alterations).
Figure 1.
Flow diagram representing numbers of patients with next-generation sequencing data available from archival tissue collected at the time of study entry, and associated outcomes data.
Univariate associations of altered genes with overall response and PFS were performed and plotted volcano-style (Supplementary Fig. S1A and S1B). Although none of the genes met the FDR threshold of 0·05, KRAS emerged as the strongest predictor across both outcomes in the binimetinib group; therefore, we chose to focus on its effects. We also investigated multivariable genomic models for PFS and overall response outcomes (Supplementary Table S3A and S3B). Notably, NRAS was the only other predictor to be independently associated with overall response in the model. As in previous reports, NRAS was mutually exclusive with KRAS in the overall cohort of 215 patients (17). Taken together with existing reports of MAPK signaling pathway mutations being associated with sensitivity to MEK inhibitors, this led us to investigate the MAPK signaling pathway as well. When defining best response by RECIST 1.1 in a binary fashion as either CR/PR or SD/PD based on KRAS mutation status, among patients treated with PCC, 33% (8/24) of patients with a KRAS mutation achieved a CR/PR compared with 19% (8/42) of patients who were KRAS WT. Among patients treated with binimetinib, 44% (20/45) of patients with a KRAS mutation achieved a CR/PR compared with 19% (17/90) of patients who were KRAS WT. Patients who harbored a KRAS mutation had 3.4 times the odds of responding to treatment with binimetinib compared with patients without a KRAS mutation [95% confidence interval (CI), 1.57–7.67; P = 0.004].
In the binimetinib treatment group, patients harboring a KRAS mutation had a 50% lower risk of a PFS event (progression or death) compared with patients without a KRAS mutation. Patients treated in the PCC group showed a smaller effect size in the same direction. Interaction between type of treatment/KRAS mutation was not significant on either additive or multiplicative scale for response (additive P = 0.8, multiplicative P = 0.5) or PFS (additive P = 0.5, multiplicative P = 0.5) outcomes (Fig. 2A).
Figure 2.
A, Binimetinib and physician's choice of chemotherapy treatment groups (n = 215), Kaplan–Meier plot of progression-free survival by treatment arm and KRAS mutation status (Mut vs. WT). B, Binimetinib and physician's choice of chemotherapy treatment groups (n = 215), Kaplan–Meier plot of progression-free survival by treatment arm and MAPK alteration status. (MEK162, binimetinib; Mut, mutant; WT, wild-type; ALT, alteration). The x-axis is percentage of patient's progression-free survival; the y-axis is time in months.
To investigate whether KRAS G12V mutations were associated with more favorable outcomes compared with other KRAS point mutations, a Cox regression was used to evaluate PFS for patients with a KRAS G12V mutation versus those with other KRAS mutations. The Fisher exact test was used to define any association between KRAS G12V (n = 19) versus other KRAS mutations (n = 26) and radiographic best response by RECIST 1.1. Among patients treated with binimetinib, there was no difference detected in the effect of KRAS G12V mutation versus other KRAS mutations on PFS or best radiographic response (PFS HR, 1.08; 95% CI, 0.45–2.59; P = 0.9; Supplementary Table S4).
In the 135 patients treated with binimetinib who had RECIST 1.1 response data available, other identified MAPK pathway alterations included the following: NRAS (n = 11, 8.1%), BRAF V600E (n = 8, 5.9%), RAF1 (n = 2, 1.5%), and NF1 (n = 7, 5.2%). In patients treated with binimetinib, the response rate (best response of CR or PR) was 41% in patients with a MAPK pathway alteration and 13% in those without a MAPK pathway alteration (Table 1). In patients treated with PCC (n = 66), the response rates were 29% and 19% in patients with and without a MAPK pathway alteration, respectively. Among patients treated with binimetinib (n = 144), longer PFS was observed in patients harboring MAPK pathway alterations compared with those without a MAPK pathway alteration (HR, 0.50; 95% CI, 0.31–0.79; P = 0.003). In patients treated with PCC (n = 71), there was a trend toward longer PFS for patients with, compared with without, a MAPK pathway alteration (HR, 0.82; 95% CI, 0.43–1.59; P = 0.6). Tests for interaction between treatment and MAPK pathway alteration were not significant on additive or multiplicative scales for both response (additive P = 0.9, multiplicative P = 0.2) and PFS (additive P = 0.4, multiplicative P = 0.3) outcomes (Fig. 2B).
Table 1.
Association between MAPK pathway-altered genes with best response by RECIST v1.1 in binimetinib-treated patients.
| Overall (n = 135) | CR/PR (n = 37; RR) | SD/PD (n = 98) | ||
|---|---|---|---|---|
| Alteration Identified | n (% altered) | n (%) | n | P a |
| KRAS: | 0.004 | |||
| ALT | 45 (33%) | 20 (44%) | 25 | |
| WT | 90 (67%) | 17 (19%) | 73 | |
| NRAS: | 0.2 | |||
| ALT | 11 (8%) | 5 (46%) | 6 | |
| WT | 124 (92%) | 32(26%) | 92 | |
| BRAFV600E: | >0.9 | |||
| ALT | 8 (6%) | 2 (25%) | 6 | |
| WT | 127 (94%) | 35(28%) | 92 | |
| RAF1: | 0.5 | |||
| ALT | 2 (1.5%) | 1 (50%) | 1 | |
| WT | 133 (99%) | 36 (27%) | 97 | |
| NF1: | >0.9 | |||
| ALT | 7 (5%) | 2 (29%) | 5 | |
| WT | 128 (95%) | 35 (27%) | 93 | |
| MAPK Pathway: | <0.001 | |||
| ALT | 68 (50%) | 28 (41%) | 40 | |
| WT | 67 (50%) | 9 (13%) | 58 |
Abbreviations: ALT, altered; RECIST, Response Evaluation Criteria in Solid Tumors; RR, response rate.
a P values calculated with the Fisher exact test.
Figure 3 displays a swimmer plot with the treatment duration and best response observed for the 144 patients treated with binimetinib. At the time of the August, 2021 data cutoff date, 5 patients remained on treatment with binimetinib. The median time to best response was 3.6 months overall and 14.6 months in the 5 patients who achieved a CR.
Figure 3.

Swimmer plot representing the duration of treatment for patients treated with binimetinib (n = 144). Best response by RECIST v1.1 criteria, reason for treatment discontinuation, and MAPK pathway alteration status are detailed in the legend.
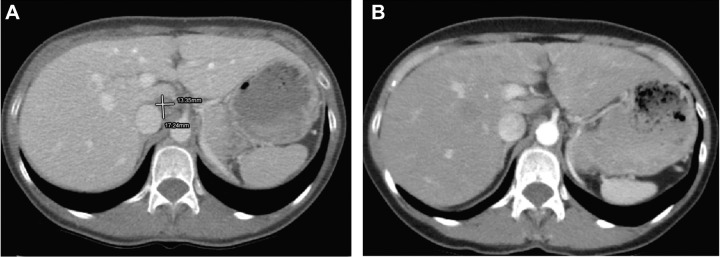
Representative CT scan images from a patient with a KRAS G12D mutation who experienced a sustained CR while on treatment with binimetinib are shown in Fig. 4. The patient was initially diagnosed with stage IIIA serous borderline tumor with non-invasive implants in 2005, at which time she underwent a total abdominal hysterectomy, bilateral salpingectomy, and surgical staging. She was diagnosed with recurrent disease with progression to LGSOC in 2013. Following exploratory laparotomy and tumor debulking, she was found to have residual disease with multiple implants on the small and large bowel mesentery. Postoperatively, she was treated with intravenous carboplatin and weekly paclitaxel plus bevacizumab, followed by 14 cycles of bevacizumab maintenance until a PET scan revealed multifocal progression of disease. A biopsy performed at the time of progression revealed a KRAS G12D mutation. In 2015, the patient was enrolled onto the MILO/ENGOT-ov11 study and initiated treatment with binimetinib. After 16 months on treatment, she achieved a CR. In 2018, she experienced a reduction in cardiac ejection fraction (EF) to 45%. Binimetinib was withheld until recovery to normal EF, at which time, the patient resumed binimetinib at a reduced dose. She had sustained CR at the time of the August, 2021 data cutoff date.
Figure 4.
CT scan images from a patient with a KRAS G12D mutation and a sustained complete response while on treatment with binimetinib. A, CT scan at study enrollment in 2015. B, CT scan in 2020 displaying sustained complete response with resolution of perihepatic adenopathy.
Discussion
Most patients with LGSOC present with advanced disease, and 70% ultimately developed recurrent or persistent disease (18). Historically, LGSOC has been both difficult to treat and challenging to study in a prospective fashion. Patients with LGSOC have clinically, histologically, and molecularly distinct disease characteristics compared with patients with high-grade disease, with a younger median age at diagnosis, lower response rates to chemotherapy, and absence of TP53 mutations (19–21). For these reasons, patients with LGSOC are generally excluded from most clinical trials evaluating novel therapies for ovarian cancer. Moreover, prospective therapeutic trials limited to patients with LGSOC have been hampered by the rarity of the disease, requiring additional time, expense, and collaboration between multiple centers for the timely accrual of patients.
The MILO/ENGOT-ov11 study has been the largest prospective phase 3 clinical trial performed in patients with histologically confirmed LGSOC, with 341 enrolled patients across 102 sites. The data presented here represent a post hoc analysis that examined the relationship between tumor mutation profile performed on FFPE tissue collected at the time of study entry as part of a pre-planned exploratory analysis and patient outcomes based on the updated August, 2021 data cutoff date.
This analysis confirms that MAPK pathway alterations, most commonly in KRAS, are frequently identified in patients with recurrent LGSOC, which is consistent with prior reports of alterations in KRAS (19%–41%), NRAS (9%–26%), and BRAF (2%–16%) in patients with LGSOC (4, 6, 22–24).
Patients who harbored a KRAS mutation were 3.4 times more likely to respond (CR or PR) to treatment with binimetinib compared with patients without a KRAS mutation (95% CI, 1.57–7.67). Among patients treated with binimetinib, 44% (20/45) of patients with a KRAS mutation achieved a CR/PR compared with 19% (17/90) of patients who were KRAS WT. Among patients treated with PCC, 33% (8/24) of patients with a KRAS mutation and 19% (8/42) of patients who were KRAS WT achieved a best response of CR or PR. Among patients treated with binimetinib, those harboring MAPK pathway alterations compared with those without such alterations had longer PFS (HR, 0.50; 95% CI, 0.31–0.79). A similar trend toward improved PFS was observed among patients harboring a MAPK pathway alteration treated with PCC (HR, 0.82; 95% CI, 0.43–1.59). Limited in vitro and in vivo data indicate that patients harboring the KRAS G12V mutation may be more sensitive to MEK inhibition compared with other KRAS variants (25–28). In our study, there was no difference in the impact of KRAS G12V mutation versus other KRAS mutations on PFS or best radiographic response. To further evaluate for any relationship between the presence of MAPK pathway alterations and clinical outcome, we examined the group of patients displaying any MAPK pathway alteration compared with those without and found that in the binimetinib-treated patients, the presence of MAPK pathway alterations was associated with longer PFS (HR, 0.50; 95% CI, 0.31–0.79). Of note, the median time to best response for the five binimetinib-treated patients who achieved a CR was 14.6 months, indicating that prolonged follow-up may be needed to ascertain patients’ true best response. This is important when considering interim analysis designs for future studies and in the clinical management of patients.
In GOG-0281, sequencing data were available for 134 patients, 44 (33%) of whom had tumors harboring activating MAPK pathway mutations in KRAS, BRAF, or NRAS. Mutations were detected in 22 (31%) of the 70 patients in the trametinib-treated group and 22 (34%) of 64 patients in the standard-of-care group (11). In the study, PFS and ORR were both markedly better in the patients treated with trametinib harboring a MAPK pathway mutation [median PFS, 13.2 months (95% CI, 9.4–20.8) vs. 7.3 months (95% CI, 5.6–12.7), respectively; ORR, 50% (95% CI, 30.2–69.8) vs. 8.3% (95% CI, 2.9–18.6), respectively]. The analysis did not find that mutation status was predictive of PFS (multiple comparison adjusted P = 0.72, test for interaction). Similar to the results presented here, in the GOG-0281 study, the ORR was more favorable with trametinib than standard-of-care therapy in patients with MAPK pathway mutations [11 (50%) of 22 vs. 2 (9%) of 22] compared with those without such mutations [4 (8%) of 45 vs. 3 (7%) of 42], although this did not reach statistical significance (multiple comparison adjusted P = 0.11, test for interaction; ref. 11).
Prior retrospective data indicate that in patients with LGSOC, the presence of a MAPK pathway alteration is associated with platinum sensitivity and prolonged survival, supporting the trend toward improved outcomes observed in patients treated with PCC in our study (14, 29). However, in patients treated with a MEK inhibitor, the presence of a MAPK pathway alteration (including KRAS) was associated with a marked difference in response, both in GOG-0281 and in the MILO/ENGOT-ov11 study. The response rate (CR/PR) for patients treated with binimetinib harboring a MAPK pathway alteration was 41% compared with 13% in patients without a MAPK pathway alteration. A lesser trend toward improved response rate was observed in patients treated with PCC (29% and 19%, respectively).
This study has several limitations. The analysis reported here is a post hoc analysis based upon sequencing of selected genes, as opposed to whole-exome or whole-genome sequencing. The low incidence of mutations and post hoc nature of the analysis limits the power to look for statistically significant associations. This analysis is primarily hypothesis generating and is limited by multiple testing. In addition, biopsy samples from patients with LGSOC frequently have low tumor content and psammomatous calcifications (14), limiting the ability to perform genetic profiling in a subset of submitted FFPE tissue specimens.
Taken together, these data show that MEK inhibitors have activity in women with LGSOC, and that mutation status should be considered when counseling patients regarding expectations of clinical benefit. Patients with recurrent LGSOC with a MAPK alteration, most commonly KRAS, were found to have higher response rates to both PCC and binimetinib treatment, with the highest response rates seen in patients who harbored a MAPK alteration and were treated with binimetinib. Given the potential for toxicity with MEK inhibitors, which can include risk of ocular adverse events, rash, edema, and congestive heart failure, a patient's molecular status may be helpful in determining sequencing of therapies and individual risk/benefit analysis. Novel therapeutics targeting the MAPK pathway are currently in development for patients with LGSOC, with new opportunities for therapeutic precision on the horizon. Future prospective studies performed in patients with LGSOC should collect data regarding patients’ somatic mutation status and consider stratification for this variable.
Conclusions
Higher response rates and longer PFS were seen in patients who harbored MAPK pathway alterations, with the highest response rates observed in patients with MAPK pathway alterations treated with binimetinib. Although interactions between treatment and KRAS/MAPK alteration status were not significant, effects were stronger and associations more significant in the binimetinib group, though the physician's choice group in particular was small in numbers. Evidence to support the hypothesis that MAPK pathway alteration is predictive of response to binimetinib would require a more adequately powered study. However, the results of this analysis provide reliable evidence that MAPK pathway alteration status has prognostic implications for patients with LGSOC. Somatic tumor testing should be routinely considered in patients with recurrent LGSOC to aid in clinical decision making, and should be accounted for in future clinical trials.
Supplementary Material
Supplementary Tables and Figures
Acknowledgments
R.N. Grisham and C. Aghajanian are funded in part by the National Cancer Institute (NCI) at the National Institutes of Health (NIH; P30 CA008748). The MILO/ENGOT-ov11 study was supported by Pfizer.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
R.N. Grisham reports personal fees from GSK, AZ, Natera, Verastem, Corcept, OncLive, PrIme, Springworks, MJH Life Sciences, and Aptitude Health outside the submitted work. I. Vergote reports grants from Oncoinvent AS, Amgen, and Roche; personal fees from Agenus, Akesobio, AstraZeneca, Bristol Myers Squibb, Deciphera Pharmaceuticals, Eisai, Elevar Therapeutics, Exelixis, F. Hoffmann-LaRoche, Genmab, GSK, Immunogen, Jazzpharma, Karyopharm, Mersana, MSD, Novocure, Novartis, Oncoinvent, OncXerna, Regeneron, Sanofi, Seagen, Sotio, Verastem Oncology, and Zentalis outside the submitted work; and travel expenses from Karyopharm, Genmab, and Novocure. S. Banerjee reports grants from AstraZeneca and GSK; personal fees from AstraZeneca, GSK, Immunogen, MSD, Mersana, Novartis, Oncxerna, Seagen, Shattuck Labs, Regeneron, Epsilogen, Novacure, Takeda, Verastem, Roche, and Pfizer outside the submitted work; and is Global Lead, ENGOTov60/GOG3052. I. Romero reports personal fees and nonfinancial support from MSD and AstraZeneca outside the submitted work; grants, personal fees, and nonfinancial support from GSK; and personal fees from Pharmamar and Roche. P. Vuylsteke reports other support from MSD and Novartis outside the submitted work, as well as grants from Roche. R.L. Coleman reports grants and personal fees from Clovis Oncology, GSK, AstraZeneca, Merck, and Genmab during the conduct of the study, as well as personal fees from Immunogen, Alkermes, Mersana, BMS, Easai, Novocure, and Zentalis outside the submitted work. F. Hilpert reports personal fees from AstraZeneca, MSD, Novartis, GSK, and Clovis during the conduct of the study. A.M. Oza reports serving on PI and Steering Committees with AstraZeneca, GSK, and Clovis as well as on Advisory Boards of AstraZeneca and Morphosys. S. Pignata reports grants and personal fees from MSD, Roche, and GSK; grants from Pfizer; and personal fees from Pharmamar outside the submitted work. C. Aghajanian reports personal fees from Roche/Genentech, Eisai/Merck, AstraZeneca/Merck, and Repare Therapeutics; grants from Abbvie, AstraZeneca, Clovis, and Roche/Genentech; and other support from GOG Foundation, Board of Directors, outside the submitted work. N. Colombo reports personal fees from Clovis, Roche, MSD, GSK, Immunogen, Mersana, Eisai, Oncxerna, and Nuvation Bio outside the submitted work. K.N. Moore reports personal fees from Aadi, AstraZeneca, Aravive, Blueprint pharma, Clovis, Caris, Duality, Eisai, GSK, Genentech/Roche, Immunogen, InxMed, Lilly, Merck, Myriad, Mersana, OncXerna, OncoNova, Novartis, Janssen, Pannavance, VBL Therapeutics, Novacure, Verastem, and Zentalis outside the submitted work; and works with AD GOG Partners (compensated), GOG Foundation (uncompensated), and ASCO BOD (uncompensated). R. Berger reports grants from Array BioPharma during the conduct of the study. C. Marth reports personal fees from Roche, Novartis, Amgen, MSD, Pharma-Mar, AstraZeneca, GSK, and Seagen outside the submitted work. D.M. O'Malley reports grants and other support from Pfizer during the conduct of the study; grants from AbbVie, Advaxis, Agenus Inc., Alkermes, Aravive Inc., Arcus Biosciences Inc., AstraZeneca, BeiGene USA Inc., Boston Biomedical, Bristol Myers Squibb, Clovis Oncology, Deciphera Pharma, Eisai, EMD Serono Inc., Exelixis, Genentech Inc., Genmab, GlaxoSmithKline, GOG Foundation, Hoffmann-La Roche Inc., ImmunoGen Inc., Incyte Corporation, Iovance Biotherapeutics, Karyopharm, Leap Therapeutics Inc., Ludwig Institute for Cancer Research, Merck & Co., Merck Sharp & Dohme Corp., Mersana Therapeutics Inc., NCI, Novartis, NovoCure, NRG Oncology, OncoC4 Inc., OncoQuest Inc., Pfizer Inc., Precision Therapeutics Inc., Prelude Therapeutics, Regeneron Pharmaceuticals Inc., RTOG, Rubius Therapeutics, Seattle Genetics (Seagen), Sutro Biopharma, SWOG, TESARO, and Verastem Inc.; and personal fees from AbbVie, AdaptImmune, Agenus Inc., Arquer Diagnostics, Arcus Biosciences Inc., AstraZeneca, Atossa Therapeutics, Boston Biomedical, Cardiff Oncology, Celcuity, Clovis Oncology, Corcept Therapeutics, Duality Bio, Eisai, Elevar, Exelixis, Genentech Inc., Genelux, GlaxoSmithKline, GOG Foundation, Hoffmann-La Roche Inc., ImmunoGen Inc., Imvax, InterVenn, Inxmed, Iovance Biotherapeutics, Janssen, Jazz Pharmaceuticals, Laekna, Leap Therapeutics Inc., Luzsana Biotechnology, Merck & Co., Merck Sharp & Dohme, Mersana Therapeutics Inc., Myriad, Novartis, NovoCure, OncoC4 Inc., Onconova, Regeneron Pharmaceuticals Inc., RepImmune, R Pharm, Roche Diagnostics, Seattle Genetics (Seagen), Sorrento, Sutro Biopharma, Tarveda Therapeutics, Toray, Trillium, Umoja, Verastem Inc., VBL Therapeutics, Vincerx Pharma, Xencor, and Zentalis outside the submitted work. A. Clamp reports grants from Array BioPharma during the conduct of the study, as well as from Advenchen, Eisai, Verastem, and Novartis outside the submitted work; grants and personal fees from AstraZeneca and MSD; and personal fees from GSK, Clovis Oncology, and Immunogen. G. Iyer reports research funding from Janssen, Mirati Therapeutics, LOXO at Lilly, Seagen, Tyra Biosciences, Aadi Bioscience, Novartis, Debiopharm Group, and Bayer; and acts in Consulting/Advisory capacity for Mirati Therapeutics, Gilead, LOXO at Lilly, Aadi Bioscience, Flare Therapeutics, and The Lynx Group. I. Ray-Coquard reports personal fees from AstraZeneca, Clovis, GSK, Deciphera, Agenus, Adaptimmun, and ESAI during the conduct of the study; grants and personal fees from Roche, BMS, and Immunogen; grants from EQRX, Daichi Sankyo, and Exelixis; and grants, personal fees, and nonfinancial support from MSD. B.J. Monk reports personal fees from Acrivon, Adaptimmune, Agenus, Akeso Bio, Amgen, Aravive, AstraZeneca, Bayer, Clovis, Easai, Elevar, EMD Merck, Genmab/Seagen, GOG Foundation, Gradalis, Heng Rui, ImmunoGen, Karyopharm, Iovance, Laekna Health Care, Merck, Mersana, Myriad, Novartis, Novocure, Onco4, Panavance, Pieris, Pfizer, Puma, Regeneron, Roche/Genentech, Sorrento, TESARO/GSK, VBL, Verastem, and Zentalis outside the submitted work. No disclosures were reported by the other authors.
Disclaimer
The funding sources had no role in the study design, in the collection, analysis and interpretation of data, in the writing of the report, or in the decision to submit the article for publication.
Authors' Contributions
R.N. Grisham: Conceptualization, data curation, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. I. Vergote: Conceptualization, data curation, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. S. Banerjee: Investigation, writing–review and editing. E. Drill: Conceptualization, data curation, formal analysis, methodology, writing–original draft, writing–review and editing. E. Kalbacher: Investigation, writing–review and editing. M.R. Mirza: Investigation, writing–review and editing. I. Romero: Investigation, writing–review and editing. P. Vuylsteke: Investigation, writing–review and editing. R.L. Coleman: Investigation, writing–review and editing. F. Hilpert: Investigation, writing–review and editing. A.M. Oza: Investigation, writing–review and editing. A. Westermann: Investigation, writing–review and editing. M.K. Oehler: Investigation, writing–review and editing. S. Pignata: Investigation, writing–review and editing. C. Aghajanian: Investigation, writing–review and editing. N. Colombo: Investigation, writing–review and editing. D. Cibula: Investigation, writing–review and editing. K.N. Moore: Investigation, writing–review and editing. J.M. del Campo: Investigation, writing–review and editing. R. Berger: Investigation, writing–review and editing. C. Marth: Investigation, writing–review and editing. J. Sehouli: Investigation, writing–review and editing. D.M. O'Malley: Investigation, writing–review and editing. C. Churruca: Investigation, writing–review and editing. G. Kristensen: Investigation, writing–review and editing. A. Clamp: Investigation, writing–review and editing. J. Farley: Investigation, writing–review and editing. G. Iyer: Data curation, formal analysis, writing–original draft, writing–review and editing. I. Ray-Coquard: Data curation, formal analysis, investigation, writing–original draft, writing–review and editing. B.J. Monk: Conceptualization, data curation, formal analysis, supervision, visualization, writing–original draft, project administration, writing–review and editing.
References
- 1. Singer G, Stöhr R, Cope L, Dehari R, Hartmann A, Cao D-F, et al. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: a mutational analysis with immunohistochemical correlation. Am J Surg Pathol 2005;29:218–24. [DOI] [PubMed] [Google Scholar]
- 2. Kurman RJ, Shih IeM. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol 2016;186:733–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smalley I, Smalley KSM. ERK inhibition: a new front in the war against MAPK pathway-driven cancers? Cancer Discov 2018;8:140–2. [DOI] [PubMed] [Google Scholar]
- 4. Wong K-K, Tsang YTM, Deavers MT, Mok SC, Zu Z, Sun C, et al. BRAF mutation is rare in advanced-stage low-grade ovarian serous carcinomas. Am J Pathol 2010;177:1611–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grisham RN, Iyer G, Garg K, DeLair D, Hyman DM, Zhou Q, et al. BRAF mutation is associated with early-stage disease and improved outcome in patients with low-grade serous ovarian cancer. Cancer 2013;119:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hunter SM, Anglesio MS, Ryland GL, Sharma R, Chiew Y-E, Rowley SM, et al. Molecular profiling of low grade serous ovarian tumours identifies novel candidate driver genes. Oncotarget 2015;6:37663–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Nieuwenhuysen E, Busschaert P, Laenen A, Moerman P, Han SN, Neven P, et al. Loss of 1p36.33 frequent in low-grade serous ovarian cancer. Neoplasia 2019;21:582–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gershenson DM, Sun CC, Bodurka D, Coleman RL, Lu KH, Sood AK, et al. Recurrent low-grade serous ovarian carcinoma is relatively chemoresistant. Gynecol Oncol 2009;114:48–52. [DOI] [PubMed] [Google Scholar]
- 9. Grisham RN, Sylvester BE, Won H, McDermott G, DeLair D, Ramirez R, et al. Extreme outlier analysis identifies occult mitogen-activated protein kinase pathway mutations in patients with low-grade serous ovarian cancer. J Clin Oncol 2015;33:4099–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farley J, Brady WE, Vathipadiekal V, Lankes HA, Coleman R, Morgan MA, et al. Selumetinib in women with recurrent low-grade serous carcinoma of the ovary or peritoneum: an open-label, single-arm, phase 2 study. Lancet Oncol 2013;14:134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gershenson DM, Miller A, Brady WE, Paul J, Carty K, Rodgers W, et al. Trametinib versus standard of care in patients with recurrent low-grade serous ovarian cancer (GOG 281/LOGS): an international, randomised, open-label, multicentre, phase 2/3 trial. Lancet 2022;399:541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Monk BJ, Grisham RN, Banerjee S, Kalbacher E, Mirza MR, Romero I, et al. MILO/ENGOT-ov11: binimetinib versus physician's choice chemotherapy in recurrent or persistent low-grade serous carcinomas of the ovary, fallopian tube, or primary peritoneum. J Clin Oncol 2020;38:3753–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Armstrong DK, Alvarez RD, Backes FJ, Bakkum-Gamez JN, Barroilhet L, Behbakht K, et al. NCCN guidelines(R) insights: ovarian cancer, version 3.2022. J Natl Compr Canc Netw 2022;20:972–80. [DOI] [PubMed] [Google Scholar]
- 14. Manning-Geist B, Gordhandas S, Liu YL, Zhou Q, Iasonos A, Da Cruz Paula A, et al. MAPK pathway genetic alterations are associated with prolonged overall survival in low-grade serous ovarian carcinoma. Clin Cancer Res 2022;28:4456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Braicu C, Buse M, Busuioc C, Drula R, Gulei D, Raduly L, et al. A comprehensive review on MAPK: a promising therapeutic target in cancer. Cancers 2019;11:1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mathur MB, VanderWeele TJ. R function for additive interaction measures. Epidemiology 2018;29:e5–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheasley D, Nigam A, Zethoven M, Hunter S, Etemadmoghadam D, Semple T, et al. Genomic analysis of low-grade serous ovarian carcinoma to identify key drivers and therapeutic vulnerabilities. J Pathol 2021;253:41–54. [DOI] [PubMed] [Google Scholar]
- 18. Gershenson DM. Low-grade serous carcinoma of the ovary or peritoneum. Ann Oncol 2016;27:i45–9. [DOI] [PubMed] [Google Scholar]
- 19. Grabowski JP, Harter P, Heitz F, Pujade-Lauraine E, Reuss A, Kristensen G, et al. Operability and chemotherapy responsiveness in advanced low-grade serous ovarian cancer. An analysis of the AGO study group metadatabase. Gynecol Oncol 2016;140:457–62. [DOI] [PubMed] [Google Scholar]
- 20. Plaxe SC. Epidemiology of low-grade serous ovarian cancer. Am J Obstet Gynecol 2008;198:459. [DOI] [PubMed] [Google Scholar]
- 21. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singer G, Oldt R, Cohen Y, Wang BG, Sidransky D, Kurman RJ, et al. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst 2003;95:484–6. [DOI] [PubMed] [Google Scholar]
- 23. Emmanuel C, Chiew Y-E, George J, Etemadmoghadam D, Anglesio MS, Sharma R, et al. Genomic classification of serous ovarian cancer with adjacent borderline differentiates RAS pathway and TP53-mutant tumors and identifies NRAS as an oncogenic driver. Clin Cancer Res 2014;20:6618–30. [DOI] [PubMed] [Google Scholar]
- 24. Chui MH, Chang JC, Zhang Y, Zehir A, Schram AM, Konner J, et al. Spectrum of BRAF mutations and gene rearrangements in ovarian serous carcinoma. JCO Precis Oncol 2021;5:PO.21.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo C, Chénard-Poirier M, Roda D, de Miguel M, Harris SJ, Candilejo IM, et al. Intermittent schedules of the oral RAF–MEK inhibitor CH5126766/VS-6766 in patients with RAS/RAF-mutant solid tumours and multiple myeloma: a single-centre, open-label, phase 1 dose-escalation and basket dose-expansion study. Lancet Oncol 2020;21:1478–88. [DOI] [PubMed] [Google Scholar]
- 26. Jänne PA, Smith I, McWalter G, Mann H, Dougherty B, Walker J, et al. Impact of KRAS codon subtypes from a randomised phase II trial of selumetinib plus docetaxel in KRAS mutant advanced non–small cell lung cancer. Br J Cancer 2015;113:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsang YT, Deavers MT, Sun CC, Kwan S‐Y, Kuo E, Malpica A, et al. KRAS (but not BRAF) mutations in ovarian serous borderline tumour are associated with recurrent low-grade serous carcinoma. J Pathol 2013;231:449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gershenson DM, Sun CC, Westin SN, Eyada M, Cobb LP, Nathan LC, et al. The genomic landscape of low-grade serous ovarian/peritoneal carcinoma and its impact on clinical outcomes. Gynecol Oncol 2022;165:560–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables and Figures
Data Availability Statement
The raw sequencing data for this study were generated at Foundation Medicine; requests for sequencing data can be sent to clinicaltrials@foundationmedicine.com. Derived data supporting the findings of this study are available from the corresponding author upon request.