Abstract
Purpose:
Human papillomavirus (HPV) is causally linked to oropharyngeal squamous cell carcinoma (OPSCC). Consensus guidelines recommend clinical exams and imaging in decreasing frequency as part of posttreatment surveillance for recurrence. Plasma tumor tissue modified viral (TTMV)-HPV DNA testing has emerged as a biomarker which can inform disease status during surveillance.
Experimental Design:
This retrospective observational cohort study involved 543 patients who completed curative-intent therapy for HPV-associated OPSCC between February 2020 and January 2022 at eight U.S. cancer care institutions. We determined the negative predictive value (NPV) of TTMV-HPV DNA for recurrence when matched to physician-reported clinical outcome data (median follow-up time: 27.9 months; range: 4.5–154).
Results:
The cohort included mostly men with a median age of 61 who had locoregionally advanced disease. HPV status was determined by p16 positivity in 87% of patients, with a positive HPV PCR/ISH among 55%; while pretreatment TTMV-HPV DNA status was unknown for most (79%) patients. Patients had a mean of 2.6 tests and almost half had three or more TTMV-HPV DNA results during surveillance. The per-test and per-patient sensitivity of the assay was 92.5% [95% confidence interval (CI): 87.5–97.5] and 87.3% (95% CI: 79.1–95.5), respectively. The NPV for the assay was 99.4% (95% CI: 98.9–99.8) and 98.4% (95% CI: 97.3–99.5), respectively.
Conclusions:
TTMV-HPV DNA surveillance testing yields few false negative results and few missed recurrences. These data could inform decisions on when to pursue reimaging following first disease restaging and could inform future surveillance practice. Additional study of how pretreatment TTMV-HPV DNA status impacts sensitivity for recurrence is needed.
Translational Relevance.
Circulating fragmented human papillomavirus (HPV) DNA is a unique tumor biomarker of HPV-driven malignancies. Tumor tissue modified viral (TTMV)-HPV DNA is associated with recurrent or residual oropharyngeal cancer when detected during posttreatment surveillance. This retrospective observational cohort study (N = 543) is the largest published observational cohort demonstrating the clinical validity of TTMV-HPV DNA correlated with deeply curated longitudinal medical records. Per-patient negative predictive value, positive predictive value, sensitivity, and specificity, were 98.4%, 94.8%, 87.3%, and 99.4%, respectively. Given per-test and per-patient specificity >99%, newly positive or rising test results strongly support further clinical evaluation for recurrence. These data suggest that posttreatment monitoring at guideline-specified routine surveillance visits (every 3–4 months in years 1–2, every 6 months in years 3–5) appears reasonable. Amassing data across multiples clinical sites and thousands of patients continues to support the use of TTMV-HPV DNA as a routine surveillance tool.
Introduction
The incidence of human papillomavirus (HPV)-driven oropharyngeal cancers continues to rise with a decrease in tobacco use observed in the United States (1). HPV, primarily subtype 16, is causally linked to most cases of oropharyngeal squamous cell carcinoma (OPSCC) arising in the tonsil or tongue base and is associated with a favorable prognosis (2). Therapy is often multimodal relying on some combination of surgery, radiation, and/or chemotherapy. After completion of therapy, the approach to surveillance is similar to other head and neck cancers, with regular fiberoptic endoscopic exams at decreasing frequency. Imaging subsequent to first restaging is generally based on the presence of symptoms or exam findings and practice patterns vary widely. In recent years, ultrasensitive plasma-based assays to detect circulating cell-free HPV tumor DNA have emerged and show promise for improving surveillance standards.
While several HPV assays have been evaluated (3), a commercially available test for tumor tissue modified viral (TTMV)-HPV DNA (NavDx, Naveris, Inc.) that identifies circulating fragmented postapoptotic tumor-associated HPV DNA has been the most studied to date (4–6). Currently, the test has been utilized in clinical practice across more than 400 sites (Naveris data on file). The potential benefits of plasma TTMV-HPV DNA for detecting disease recurrence were demonstrated in two recent studies. Chera and colleagues reported a 94% positive predictive value (PPV) for two consecutively positive test results with a median lead-time to biopsy-proven diagnosis of 4 months in a small prospective trial of patients with HPV-associated OPSCC treated with chemoradiation (6). This was followed by a large retrospective clinical case series of more than a thousand patients tested ≥3 months from completion of curative-intent therapy pooled across more than a hundred institutions which corroborated a PPV of 95% for identifying occult disease recurrence but requiring only a single positive test (4). Furthermore, detectable HPV circulating tumor DNA following surgery for OPSCC negatively impacts recurrence-free survival (7), even when identified on postoperative day 1 following resection (8).
Building on this prior work, we report the results of a multi-institutional, real-world observational cohort study that evaluates the negative predictive value (NPV) of plasma TTMV-HPV DNA obtained during surveillance among HPV-associated OPSCC survivors. We incorporated detailed physician-reported clinical follow-up and longitudinal outcome data to further understand the role of plasma TTMV-HPV DNA monitoring as part of posttreatment surveillance.
Materials and Methods
Study participants and oversight
Patients diagnosed and treated for primary HPV-driven OPSCC without known distant metastatic disease were included if they underwent plasma TTMV-HPV DNA testing 3 or more months following completion of initial curative-intent treatment (any modality) between February 2020 and January 2022. A 3-month cut-off point was chosen based on known clearance kinetics of TTMV-HPV DNA from circulation (5), allowing transient treatment-related signal to resolve. Tumor HPV status was reported by the treating clinician using the surrogate marker p16 by IHC, direct HPV tissue detection by ISH or PCR, and/or with a detectable plasma TTMV-HPV DNA test result. Pretreatment and on-treatment plasma TTMV-HPV DNA results were recorded, if available. Eligible patients had to have a minimum of one TTMV-HPV DNA test result obtained during routine surveillance, with serial testing intervals dictated by the ordering clinician. Institutional Review Board (IRB) and data sharing approval was obtained at each participating site. A waiver of written informed consent was also granted by a central, independent IRB (WCG IRB; sponsor protocol number: NAV11042020), in line with recognized ethical guidelines, considering the limited risk associated with data collection.
Clinical data annotation
Trained investigators at participating sites performed in-depth retrospective chart review to identify eligible patients. Patients were assigned a unique study identifier and data were electronically submitted to compile a central deidentified database that included: age, sex as a biological variable, race, ethnicity, smoking status, date of initial diagnosis, location of the primary tumor, clinical and/or pathologic tumor staging [American Joint Committee on Cancer staging manual 2017, 8th edition (9)], and method of HPV confirmation. Treatment history (start and end dates of curative-intent and salvage therapy), imaging history [date, modality (CT, MRI, and/or PET-CT), result (active disease, indeterminate or suspicious, or no evidence of disease)], biopsy history [date, location, HPV status, method of HPV confirmation, result (negative for cancer, indeterminate or suspicious, positive for cancer of the oropharynx, positive for non-oropharynx cancer)], clinical visit history [date, clinical status (active disease, indeterminate or suspicious, no evidence of disease) determined by exam and fiberoptic endoscopy], and outcomes (alive or deceased, date of death or next follow-up, surveillance modality planned for next follow-up) were all recorded.
TTMV-HPV DNA testing
Test results included TTMV-HPV DNA score, HPV subtype, and the clinician reported interval between the test(s) and the end of treatment (categorized as: 3–6 months, 6–12 months, 12–24 months, or >24 months posttreatment). TTMV-HPV DNA was measured and fragment profile analyzed using the commercially available assay as described previously (5, 6, 10). TTMV-HPV DNA uses droplet digital PCR coupled with algorithmic analysis of tumor-specific HPV DNA fragmentation patterns. This approach detects tumor-derived HPV DNA from five high-risk HPV subtypes (16, 18, 31, 33, and 35). Results are reported as the TTMV-HPV DNA score, which reflects the normalized number of TTMV-HPV DNA fragments per milliliter of plasma. Scores are categorized as positive, indeterminate, or negative. Scores >7 (for HPV subtype 16) or >12 (for HPV subtypes 18, 31, 33, or 35) were considered positive, scores between 5 and 7 (HPV 16) or 5 and 12 (HPV 18, 31, 33, or 35) were considered indeterminate, while scores <5 were considered negative, regardless of HPV subtype.
Data validation
The participating clinical investigators were responsible for validation of their respective site data. Central validation of the data was performed by a validator blinded to data collection, with any discrepancies reviewed with the respective site investigator.
Statistical analysis
Data were summarized descriptively, and assay performance metrics were calculated to determine the accuracy of surveillance test predictions. No formal power analysis or randomization was employed. NPV was defined as the probability that a patient with a negative TTMV-HPV DNA score has no evidence of clinically identifiable disease at or within 3 months of testing. Time to event endpoints were summarized using the Kaplan–Meier method with recurrence-free and overall survival defined from the date of diagnosis to first confirmed recurrence or death, respectively, with censoring at last follow-up when appropriate. All statistical tests were two sided and a P < 0.05 was considered significant.
Data sharing statement
The data generated in this study comprised clinically annotated medical records by treating investigators. While these data are not publicly available to ensure patient privacy, they are available upon reasonable request from the corresponding author.
Results
Study population
Eight institutions across the United States participated with 543 patients included. Each site contributed a median of 57 patients (range: 21–194). Patient characteristics were typical of an HPV-positive OPSCC population with mostly men (466, 86%) who identified as Caucasian or White (471, 87%) and a median age of 61 (range: 26–86; Table 1). More than half of patients were never smokers. Most had T0-2 (423, 78%) and N0-2 (531, 98%) disease at initial staging, and 308 (57%) received concurrent chemoradiation as part of initial treatment. Most had HPV association determined by p16 IHC positivity (472, 87%) with 229 (42%) also positive by HPV ISH or PCR testing. Furthermore, 71 (13%) patients had positive HPV ISH or PCR testing without known p16 status.
Table 1.
Patient characteristics.
| Characteristic | N = 543 (%) |
|---|---|
| Age, years (median, range) | 61 (26–86) |
| Sex | |
| Male | 466 (86) |
| Female | 77 (14) |
| Race and ethnicity | |
| White | 471 (87) |
| Black or African American | 15 (3) |
| Asian | 6 (1) |
| Hispanic | 4 (1) |
| Native Hawaiian or other Pacific Islander | 1 (<1) |
| Preferred not to answer | 46 (9) |
| Smoking status | |
| Never | 291 (54) |
| Former (≥10 pack-years) | 230 (42) |
| Current | 22 (4) |
| Site of primary disease | |
| Tonsil | 267 (49) |
| Base of tongue | 223 (41) |
| Overlapping sites (tonsil/base of tongue) | 23 (4) |
| Unknown primary of the head and neck | 30 (6) |
| HPV testing methoda | |
| p16+ (by IHC) only | 242 (45) |
| p16+ and PCR or HPV ISH+ | 229 (42) |
| PCR or HPV ISH+ only (p16 unknown) | 71 (13) |
| p16+ and PCR or ISH− | 1 (<1) |
| HPV subtypeb | |
| 16 | 151 (86) |
| 18 | 2 (1) |
| 31 | 2 (1) |
| 33 | 11 (6) |
| 35 | 9 (5) |
| Initial primary tumor stagingc | |
| T0 | 25 (5) |
| T1 | 184 (34) |
| T2 | 214 (39) |
| T3 | 82 (15) |
| T4 | 37 (7) |
| Unknown | 2 (<1) |
| Nodal status at baselinec | |
| N0 | 58 (11) |
| N1 | 367 (68) |
| N2 | 106 (19) |
| N3 | 11 (2) |
| Unknown | 2 (<1) |
| Initial treatment modality | |
| Surgery | 121 (22) |
| Chemoradiation | 227 (42) |
| Surgery + RT | 84 (15) |
| Surgery + chemoradiation | 81 (15) |
| Other (chemotherapy, radiation only, immunotherapy, or EGFR inhibitor) | 30 (6) |
| Number of patients with pretreatment or baseline testing results available | 112 (21) |
Abbreviations: EGFR, epidermal growth factor receptor; HPV, human papillomavirus; IHC, immunohistochemistry; ISH, in situ hybridization; PCR, polymerase chain reaction; RT, radiotherapy.
aNo patients were p16 negative and confirmatory HPV ISH or PCR positive.
bHPV status was only determined for those with at least one positive or detectable test result, so the total number is equal to 175.
cAccording to American Joint Committee on Cancer staging manual 2017, 8th edition staging (if both pathologic and clinical staging were available, pathologic stage is reported).
TTMV-HPV DNA testing patterns
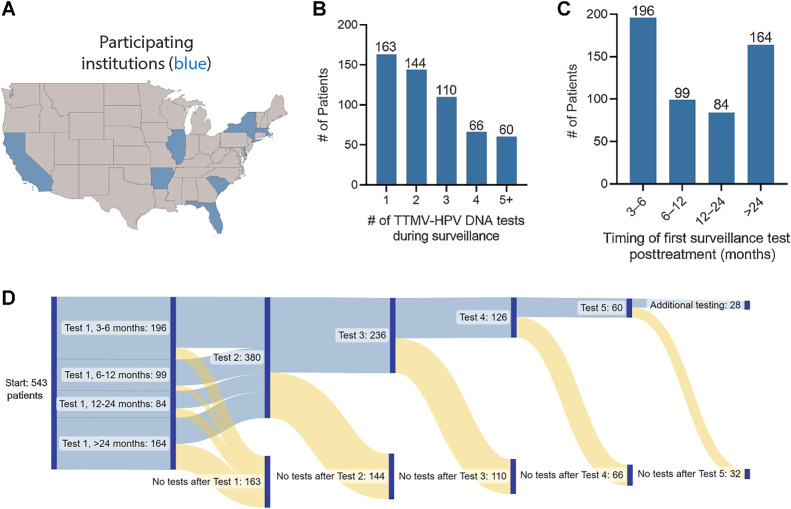
Given the recent introduction of the test into clinical practice (March 2020), baseline or pretreatment TTMV-HPV DNA testing was not required for inclusion in this surveillance study but was available for 112 (21%) patients. Most patients had >1 TTMV-HPV DNA test performed ≥3 months posttreatment (380, 70%) with 236 (44%) having three or more test results during surveillance (Fig. 1A–C). The first of these tests was often performed within the first 3–6 months after completion of therapy (196, 36%) with 164 (30%) having a test result >24 months after completing treatment. The median time to tests collected between 0–3, 3–6, 6–12, 12–24, and >24 months posttreatment was 1.13 (range: 0.03–2.97), 3.90 (range: 3.00–5.97), 8.78 (range: 6.00–11.97), 16.47 (range: 12.00–23.97), and 38.65 (range: 24.03–133.23), respectively.
Figure 1.
TTMV-HPV DNA surveillance testing patterns among HPV-associated oropharyngeal cancer survivors. A, U.S. geographic map plotting states where patient samples were collected from participating institutions. B, Bar graph plotting the number of TTMV-HPV DNA test results among 543 total patients in the cohort. C, Bar graph plotting the time interval at which test results were obtained following the completion of curative-intent treatment among 543 total patients. D, Sankey diagram integrating the number of test results per patient and the time intervals at which testing occurred from left to right.
Pretreatment TTMV-HPV DNA
Pretreatment (or baseline) TTMV-HPV DNA was detectable in 96 of 112 (86%) patients with available test results at baseline, with a median score of 337 (range: 8–65, 458) and the most common subtype being HPV16 (88%) followed by HPV33 (6%), HPV35 (4%), and HPV31 (1%). Of the 96 patients with detectable pretreatment scores, 16 (17%) had their first negative result during treatment, 58 (61%) were negative by month 3 posttreatment, 19 (20%) were first negative at 3 to 6 months posttreatment, and 2 had their first negative value at 12 to 24 months posttreatment (but in each case, it was their first posttreatment result). A single patient remained detectable from baseline following curative-intent therapy (HPV16 score 152 at baseline, and 28 days after treatment ended their score was 30; imaging at 38 days confirmed locally persistent disease).
Sensitivity and NPV of TTMV-HPV DNA
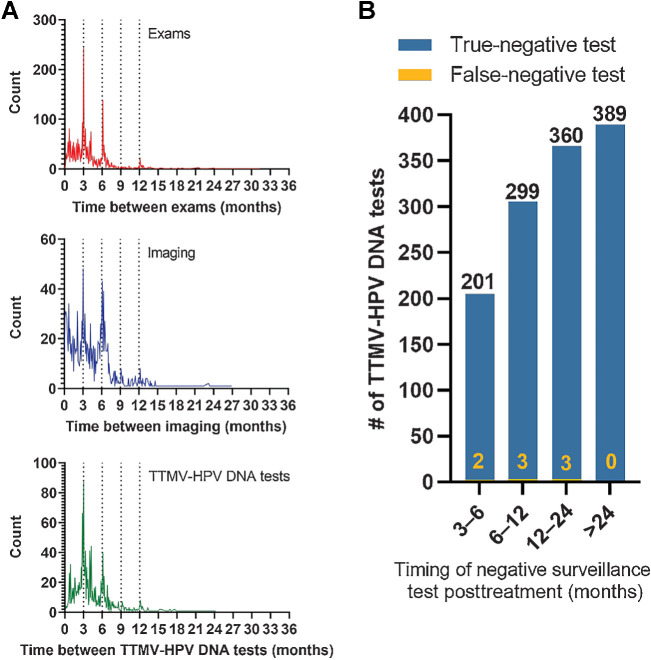
There were 55 patients with confirmed recurrences in this population cohort (10%). To determine NPV, we aligned patient-level clinical data documenting clinical status [active disease or negative (either clinically indeterminate or no evidence of disease)] at the time of each negative test result date (Fig. 2A). Results of the most time-relevant clinical exam or imaging were assigned to each TTMV-HPV DNA test. We defined two levels of relevance from greatest to least. In the absence of the most relevant result, the next most relevant was used instead: (i) an exam or imaging performed the same day the test was collected; (ii) the most recent prior exam or imaging prior to the test collection date. Negative and indeterminate test results whose most relevant clinical status was indeterminate were considered false negatives if recurrence was detected <3 months after the test date (Fig. 2B). By aligning clinician-reported disease status from exams and/or imaging with testing results, we observed an overall per-patient sensitivity of 87.3% [95% confidence interval (CI): 79.1–95.5] and NPV of 98.4% (95% CI: 97.3–99.5) for the assay among the entire study cohort (Table 2). These metrics were similar when including only the patients with a pretreatment positive TTMV-HPV DNA score. Notably, 222 test results were performed on the same day as imaging with only four discordant results (<2%). Furthermore, we report accuracy metrics for all imaging studies performed in follow-up among the cohort (Table 3). Metrics were similarly calculated for imaging as for the TTMV-HPV DNA assay. The per-patient sensitivity and NPV of imaging was 85.5% (95% CI: 77.6–93.4) and 97.4% (95% CI: 95.9–98.9), respectively.
Figure 2.
NPV of TTMV-HPV DNA surveillance testing among HPV-associated oropharyngeal cancer survivors. A, Histogram plots of ordering patterns of clinical exams, imaging, and TTMV-HPV DNA tests during surveillance (top to bottom). B, Bar graph showing all true and false negative TTMV-HPV DNA results (arranged by timing of the test in relation to posttreatment surveillance follow-up intervals, x-axis).
Table 2.
Summary of test performance measures.
| Per-test accuracy measures | |||
|---|---|---|---|
| Disease “+” | Disease “−” | Predictive valuea | |
| Test positive | 99 | 5 | PPV = 95.2% (91.1–99.3) |
| Test negative | 8 | 1,264 | NPV = 99.4% (98.9–99.8) |
| Sensitivity and Specificity | Sn = 92.5% (87.5–97.5) | Sp = 99.6% (99.3–99.9) | |
| Per-patient accuracy measures | |||
| Test positive | 55 | 3c | PPV = 94.8% (89.1–100) |
| Test negative | 8b | 495 | NPV = 98.4% (97.3–99.5) |
| Sensitivity and Specificity | Sn = 87.3% (79.1–95.5) | Sp = 99.4% (98.7–100) | |
Note: 95% CI shown in parentheses for respective values.
Abbreviations: NPV, negative predictive value; PPV, positive predictive value; Sn, sensitivity; Sp, specificity.
aOne patient was censored from the PPV calculation as they were non-compliant with follow-up despite a TTMV-HPV DNA score of >800 and subsequently died.
bOnly 2 of the 8 patients with a false negative test had pretreatment or baseline testing results. Therefore, baseline detectability of TTMV-HPV DNA was not confirmed among 6 of the 8 patients.
cIndeterminate test results that prompted clinical action via a biopsy between 1 and 60 days later were considered positive test results, whereas indeterminate results that did not prompt clinical action were considered negative.
Table 3.
Summary of imaging performance measures.
| Per-scan accuracy measures | |||
|---|---|---|---|
| Disease “+” | Disease “−” | Predictive valuea | |
| Positiveb | 92 | 30 | PPV = 75.4% (67.8–83.1) |
| Negative | 36 | 1,721 | NPV = 98.0% (97.3–98.6) |
| Sensitivity and Specificity | Sn = 71.8% (64.1–79.7) | Sp = 98.3% (97.7–98.8) | |
| Per-patient accuracy measures | |||
| Positive | 65 | 27b | PPV = 70.7% (61.3–80.0) |
| Negative | 11 | 419 | NPV = 97.4% (95.9–98.9) |
| Sensitivity and Specificity | Sn = 85.5% (77.6–93.4) | Sp = 93.9% (91.7–96.2) | |
Note: 95% CI shown in parentheses for respective values.
Abbreviations: NPV, negative predictive value; PPV, positive predictive value; Sn, sensitivity; Sp, specificity.
aOne patient was censored from the PPV calculation as they were non-compliant with follow-up despite a TTMV-HPV DNA score of >800 and subsequently died.
bIndeterminate results that prompted clinical action via a biopsy between 1 and 60 days later were considered positive scans, whereas indeterminate results that did not prompt clinical action were considered negative.
The median duration of surveillance follow-up measured from the end of curative-intent treatment and censored at the time of last recorded follow-up was 27.9 months (range: 4.5–154). Of 543 patients in the study cohort, 516 (95%) had one or more negative test result ≥3 months posttreatment. The total number of negative test results across all patients was 1,257 yielding a per-test NPV of 99.3% (95% CI: 98.9–99.8) and per-test sensitivity of 92.5% (95% CI: 87.5–97.5); of these, 203 (16%) were collected at 3 to 6 months, 302 (24%) at 6 to 12 months, 363 (29%) at 12 to 24 months, and 389 (31%) at >24 months posttreatment. The median time from one negative surveillance test to the next test was 139 days (or 4.6 months) and the median time from a negative test to the patient's next surveillance event (defined as any clinical follow-up, including exams, imaging, biopsies, treatments, or another TTMV-HPV DNA test) was 95 days (3.2 months).
Eight false negative test results were noted among 8 unique patients [per-patient sensitivity of 87.3% (95% CI: 79.1–95.5)]; 2 were collected 3 to 6 months, 3 were collected 6 to 12 months, and 3 were collected at 12 to 24 months posttreatment. Two (25%) had known baseline TTMV-HPV DNA testing which was positive (score range: 23–38); the other 6 did not have baseline values reported. The HPV tumor status of each recurrence was confirmed by p16 IHC (6/8) and/or HPV PCR (2/8). Four had locoregional disease recurrence and four had pulmonary or mediastinal metastatic disease at the time of the false negative result (patient examples shown in Supplementary Fig. S1). Notably, two of the eight results were obtained in patients who were on salvage therapy (had already recurred and were on secondary surveillance).
Specificity and PPV of TTMV-HPV DNA
A prior report of TTMV-HPV DNA obtained among 1,076 patients in a large retrospective case series yielded 80 patients with positive results, with a reported PPV of 95% (76/80; ref. 4), which has now increased to 98% (78/80) with more mature follow-up. In the current cohort, 53 of 543 (10%) patients had one or more positive TTMV-HPV DNA tests during surveillance (most were HPV16 positive). Of these, 43 had a positive test associated with a single recurrence, and 6 had multiple instances where a positive test coincided with recurrence for a total of 55 recurrences with an associated positive test. Seventeen (31%) had clinically active disease at the time of testing, while 38 (69%) had no clinically identifiable disease at the time of testing but later recurred yielding a per-patient PPV of 94.8% (95% CI: 89.1–100) and per-patient specificity of 99.4% [95% CI: 98.7–100; per-test PPV: 95.2% (95% CI: 91.1–99.3); per-test specificity: 99.6% (95% CI: 99.3–99.9)]. Of the 222 tests that were drawn on the same day as imaging, 7 were positive while imaging was negative or equivocal, but all later developed biopsy-proven recurrence. We additionally report accuracy metrics for all imaging studies performed in follow-up among the cohort (Table 3). The per-patient specificity and PPV of imaging was 93.9% (95% CI: 91.7–96.2) and 70.7% (95% CI: 61.3–80.0), respectively.
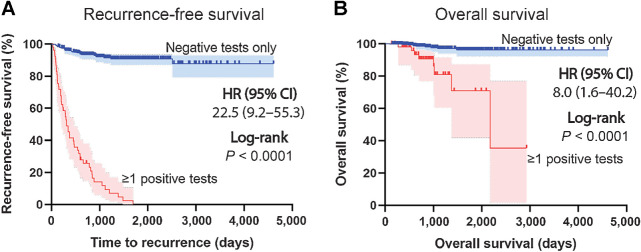
The median follow-up time was 28.7 months (range: 4.5–97.6), median time from a negative to positive result was 91 days (range: 39–189), and median lead-time from a positive test to confirmation of recurrence was 53 days (range: 7–610). Four of the 53 (8%) had a positive test but had not been diagnosed with recurrence at last follow-up. Of the 38 recurrences for which location information was available, 24 (63%) were locoregional and 14 (37%) were to distant sites (primarily the lungs). Both recurrence-free and overall survival were significantly worse for those patients with any positive TTMV-HPV DNA test result identified during surveillance, respectively [HR, 22.5 (95% CI: 9.2–55.3), P < 0.0001; HR, 8.0 (95% CI: 1.6–40.2), P < 0.0001; Fig. 3A and B].
Figure 3.
Survival among HPV-associated oropharyngeal cancer survivors based on surveillance TTMV-HPV DNA test results. Recurrence-free survival (A) and overall survival (B) curves separated by patients in the entire study cohort with negative surveillance TTMV-HPV DNA results vs. those with a positive TTMV-HPV DNA test result at any timepoint during surveillance; Kaplan–Meier method; log-rank testing (two-sided).
Discussion
Our findings continue to support the clinical utility of TTMV-HPV DNA as a circulating tumor-derived biomarker to monitor HPV-driven oropharyngeal carcinoma survivors for recurrence during posttreatment surveillance. Our initial report of a 95% PPV for TTMV-HPV DNA in identifying occult disease recurrence was published in 2022 from a large, multi-institutional observational cohort (4). The current study is unique in several ways. We aimed to capture pretreatment testing values and clinical parameters at baseline when available, and our median follow-up time was extended to 2 years. We again engaged multiple U.S. institutions with most sites not represented in our prior report (and 40% of patients included in this cohort were newly added), and we required clinicians at each site to record clinical and endoscopic exam findings, imaging, and/or biopsy reports in follow-up to confirm disease status around testing. The latter was critical to adequately estimate accuracy measurements.
With an overall sensitivity (per-patient) of 87.3% and NPV (per-patient) of 98.4% at a median of 2 years of follow-up, we demonstrate that the TTMV-HPV DNA test resulted in few false negative values and few cases of disease missed. Specifically, 8 of 56 patients with active disease had a negative test value (false negative). However, only 2 of these 8 patients were known to have a baseline positive TTMV-HPV DNA. It is possible the other 6 were never detectable pretreatment (which may occur up to 10% of the time) in which case the utility of using the test in surveillance would be limited. This is a valuable point as it supports using the test for surveillance monitoring among patients with a known positive TTMV-HPV DNA result preceding a negative result. It is known that patients with undetectable pretreatment plasma TTMV-HPV DNA more often have an N0 clinical stage, and that larger and more avid lymph nodes predict initial detectability and higher scores at baseline (11). In such cases, it is important to confirm both the presence of HPV in primary tumor tissue by direct methods and to determine the pretreatment TTMV-HPV DNA level to determine the utility of following plasma TTMV-HPV DNA testing during surveillance. Prospective data to support the use of regular surveillance imaging after first restaging for head and neck cancer survivors are lacking. Our findings suggest TTMV-HPV DNA could be sequentially used as part of surveillance to mitigate the need for unnecessary imaging exams and the need to evaluate incidental findings—as available series have shown that PET-CT has a similar NPV to TTMV-HPV DNA testing (12–15). Furthermore, the per-patient NPV and sensitivity of imaging and the assay were similar in the current study, while the PPV and specificity of TTMV-HPV DNA were higher. If using the assay results in a reduction in imaging utilization, it has also been projected that it will have a cost-saving impact (15). Similarly, the frequency of clinical exams with flexible endoscopy among asymptomatic patients may be decreased given the large number of exams required to detect a single recurrence (16).
These data corroborate our prior report of high PPV for the TTMV-HPV DNA assay, with 92% of patients in this study with a positive assay (both per-test and per-patient) having active or occult disease. Given the generally favorable prognosis of HPV-positive oropharyngeal cancer, only 58 were noted to have active disease or recurrence with relevant testing among >500 patients in the current cohort which impacts estimation of PPV. However, the per-patient and per-test specificity of >99% suggests very few false positive test results are expected. Therefore, in the face of a newly detectable or rising TTMV-HPV DNA identified during surveillance the authors continue to strongly support the use of exam and imaging (preferably a PET-CT) to evaluate for disease recurrence. A potential strength of the assay may be to lower the threshold for minimal residual disease detection which could have implications for early initiation of therapy to improve patient outcomes.
We demonstrate that most clinicians in practice use the assay at routine surveillance intervals as recommended by the National Comprehensive Cancer Network (NCCN) guidelines, obtained when patients return for clinical and fiberoptic examination. Nonetheless, the median lead-time from a positive test to confirmation of recurrence was still nearly 2 months. While the optimal timing and frequency of TTMV-HPV DNA testing is not defined, these data would suggest monitoring at routine surveillance visits [every 3–4 months in years 1–2, every 6 months in years 3–5 as recommended by the NCCN guidelines (17)] is reasonable.
These findings further clarify the potential clinical utility of TTMV-HPV DNA as a blood-based tumor biomarker that can be incorporated into clinical practice for HPV OPSCC disease–related surveillance. The authors acknowledge the limitations of retrospective observational studies, but these real-world data reflect an important geographically diverse, multi-institutional cohort with a large sample representative of the broader HPV-positive oropharyngeal cancer population across the United States. A prospective surveillance study using the TTMV-HPV DNA assay to confirm occult disease recurrence detection and guide imaging use has recently completed accrual (NCT04965792). The amassing data among thousands of patients across several studies continues to support the use of TTMV-HPV DNA monitoring as part of routine HPV-related disease surveillance.
Supplementary Material
Supplementary Figure S1: Case examples of false negative TTMV-HPV DNA results identified during surveillance
Acknowledgment
This work was not supported by any specific funding outside of data and analytic support from Naveris.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
G.J. Hanna reports personal fees from Naveris during the conduct of the study. J. Jabalee reports other support from Naveris, Inc. during the conduct of the study, as well as other support from Naveris, Inc. outside the submitted work. E.M. Rettig reports non-financial support from Naveris outside the submitted work. E. Thrash reports personal fees from Naveris during the conduct of the study. N.S. Kalman reports personal fees from Naveris outside the submitted work.A. Raben reports personal fees from Bristol Myers Squibb and other support from Castle Bioscience outside the submitted work. J.M. Kaczmar reports grants from Naveris during the conduct of the study. P. Gupta reports personal fees and other support from Naveris, Inc during the conduct of the study. C. Kuperwasser reports personal fees and non-financial support from Naveris Inc during the conduct of the study. C. Del Vecchio Fitz reports personal fees from Naveris, Inc. during the conduct of the study. B.M. Berger reports personal fees and other support from Naveris, Inc. during the conduct of the study, as well as personal fees and other support from Naveris, Inc outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
G.J. Hanna: Conceptualization, data curation, formal analysis, supervision, investigation, methodology, writing–original draft, writing–review and editing. S.A. Roof: Conceptualization, data curation, investigation, methodology, writing–original draft, writing–review and editing. J. Jabalee: Conceptualization, resources, data curation, software, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. E.M. Rettig: Data curation, investigation, writing–original draft, writing–review and editing. R. Ferrandino II: Data curation, investigation, writing–review and editing. S. Chen: Data curation, investigation, writing–review and editing. M.R. Posner: Data curation, investigation, writing–review and editing. K.J. Misiukiewicz: Data curation, investigation, writing–review and editing. E.M. Genden: Data curation, investigation, writing–review and editing. R.L. Chai: Data curation, investigation, writing–review and editing. J. Sims: Data curation, investigation, writing–review and editing. E. Thrash: Data curation, investigation. S.J. Stern: Data curation, investigation, writing–review and editing. N.S. Kalman: Data curation, investigation, writing–review and editing. S. Yarlagadda: Data curation, investigation, writing–review and editing. A. Raben: Data curation, investigation, writing–review and editing. L. Clements: Data curation, investigation, writing–review and editing. A. Mendelsohn: Data curation, investigation, writing–review and editing. J.M. Kaczmar: Data curation, investigation, writing–review and editing. Y. Pandey: Data curation, investigation, writing–review and editing. M. Bhayani: Data curation, investigation, writing–review and editing. P. Gupta: Conceptualization, resources, supervision, funding acquisition, methodology, project administration, writing–review and editing. C. Kuperwasser: Conceptualization, resources, data curation, supervision, validation, investigation, methodology, writing–review and editing. C. Del Vecchio Fitz: Conceptualization, resources, data curation, software, formal analysis, supervision, validation, investigation, methodology, writing–original draft. B.M. Berger: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, methodology, writing–original draft, project administration.
References
- 1. Vokes EE, Agrawal N, Seiwert TY. HPV-associated head and neck cancer. J Natl Cancer Inst 2015;107:djv344. [DOI] [PubMed] [Google Scholar]
- 2. Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 2011;29:4294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xie DX, Kut C, Quon H, Seiwert TY, D'Souza G, Fakhry C. Clinical uncertainties of circulating tumor DNA in human papillomavirus-related oropharyngeal squamous cell carcinoma in the absence of National Comprehensive Cancer Network guidelines. J Clin Oncol 2023;41:2483–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berger BM, Hanna GJ, Posner MR, Genden EM, Lautersztain J, Naber SP, et al. Detection of occult recurrence using circulating tumor tissue modified viral HPV DNA among patients treated for HPV-driven oropharyngeal carcinoma. Clin Cancer Res 2022;28:4292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chera BS, Amdur RJ, Green R, Shen C, Gupta G, Tan X, et al. Phase II trial of de-intensified chemoradiotherapy for human papillomavirus-associated oropharyngeal squamous cell carcinoma. J Clin Oncol 2019;37:2661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chera BS, Kumar S, Shen C, Amdur R, Dagan R, Green R, et al. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV-associated oropharyngeal cancer. J Clin Oncol 2020;38:1050–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Routman DM, Kumar S, Chera BS, Jethwa KR, Van Abel KM, Frechette K, et al. Detectable postoperative circulating tumor human papillomavirus DNA and association with recurrence in patients with HPV-associated oropharyngeal squamous cell carcinoma. Int J Radiat Oncol Biol Phys 2022;113:530–8. [DOI] [PubMed] [Google Scholar]
- 8. O'Boyle CJ, Siravegna G, Varmeh S, Queenan N, Michel A, Pang KCS, et al. Cell-free human papillomavirus DNA kinetics after surgery for human papillomavirus-associated oropharyngeal cancer. Cancer 2022;128:2193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O'Sullivan B, Huang SH, Su J, Garden AS, Sturgis EM, Dahlstrom K, et al. , Development and validation of a staging system for HPV-related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON-S): a multicentre cohort study. Lancet Oncol 2016;17:440–51. [DOI] [PubMed] [Google Scholar]
- 10. Gunning A, Kumar S, Williams CK, Berger BM, Naber SP, Gupta PB, et al. Analytical validation of NavDx, a cfDNA-based fragmentomic profiling assay for HPV-driven cancers. Diagnostics 2023;13:725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rettig EM, Wang AA, Tran NA, Carey E, Dey T, Schoenfeld JD, et al. Association of pretreatment circulating tumor tissue-modified viral HPV DNA with clinicopathologic factors in HPV-positive oropharyngeal cancer. JAMA Otolaryngol Head Neck Surg 2022;148:1120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ho AS, Tsao GJ, Chen FW, Shen T, Kaplan MJ, Colevas AD, et al. Impact of positron emission tomography/computed tomography surveillance at 12 and 24 months for detecting head and neck cancer recurrence. Cancer 2013;119:1349–56. [DOI] [PubMed] [Google Scholar]
- 13. Vainshtein JM, Spector ME, Stenmark MH, Bradford CR, Wolf GT, Worden FP, et al. Reliability of post-chemoradiotherapy F-18-FDG PET/CT for prediction of locoregional failure in human papillomavirus-associated oropharyngeal cancer. Oral Oncol 2014;50:234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Canavan JF, Harr BA, Bodmann JW, Reddy CA, Ferrini JR, Ives DI, et al. Impact of routine surveillance imaging on detecting recurrence in human papillomavirus associated oropharyngeal cancer. Oral Oncol 2020;103:104585. [DOI] [PubMed] [Google Scholar]
- 15. Ward MC, Miller JA, Walker GV, Moeller BJ, Koyfman SA, Shah C. The economic impact of circulating tumor-tissue modified HPV DNA for the post-treatment surveillance of HPV-driven oropharyngeal cancer: a simulation. Oral Oncol 2022;126:105721. [DOI] [PubMed] [Google Scholar]
- 16. Masroor F, Corpman D, Carpenter DM, Ritterman Weintraub M, Cheung KHN, Wang KH. Association of NCCN-recommended posttreatment surveillance with outcomes in patients with HPV-associated oropharyngeal squamous cell carcinoma. JAMA Otolaryngol Head Neck Surg 2019;145:903–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. National Comprehensive Cancer Network. Head and Neck Cancers (Version 1.2023). Available from: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1437. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Case examples of false negative TTMV-HPV DNA results identified during surveillance