Fig. 3.
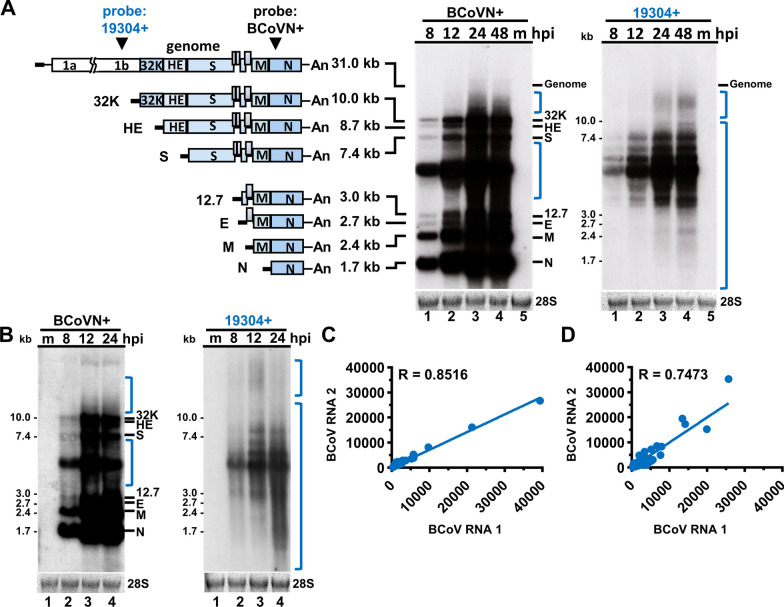
Noncanonical coronavirus transcripts are experimentally validated and reproducible. A Left panel: Diagram depicting the BCoV genome and canonical sgmRNAs. Middle and right panels: Detection of noncanonical transcripts by Northern blotting. HRT-18 cells were infected with BCoV at an MOI of 0.1, and total cellular RNA was collected at 8, 12, 24 and 48 h postinfection (hpi). In addition to the genome and canonical sgmRNAs (denoted by black bars, middle panel), signals representing noncanonical transcripts (denoted by blue brackets, middle and right panels) were detected by Northern blotting with the 32P-labeled primer probes BCoVN + (middle panel) and 19,304 + (right panel). B Reproducibility of noncanonical transcripts (denoted by blue brackets) detected by Northern blotting. C–D Reproducibility of canonical transcripts (C) and noncanonical transcripts (D) from two biological replicates. The dots in (C) and (D) correspond to the read numbers of certain noncanonical transcript species measured by nanopore direct RNA sequencing with RNA samples (RNA 1 and RNA2) collected from two independent experiments of virus infection. The noncanonical transcript species with a read count of ≥ 5 were selected from RNA samples: RNA1 and RNA2, and the dot was mapped according to the reads measured from RNA1 (X-axis) and RNA 2 (Y-axis). The reproducibility was measured in reads per kilobase per million mapped sequence reads (RPKM) and evaluated by Spearman’s correlation coefficient. R = 0.000–0.3999, low reproducibility; R = 0.4000–0.5999, moderate reproducibility; R = 0.6000–1.0000, high reproducibility. kb, kilobase; m, mock-infected cells; hpi, hours postinfection. 28S, 28S rRNA; An, poly(A) tail; 32 K, 32 kDa protein; HE, hemagglutinin/esterase; S, spike protein; 12.7, 12.7 kDa protein; E, envelope protein; M, membrane protein; N, nucleocapsid protein