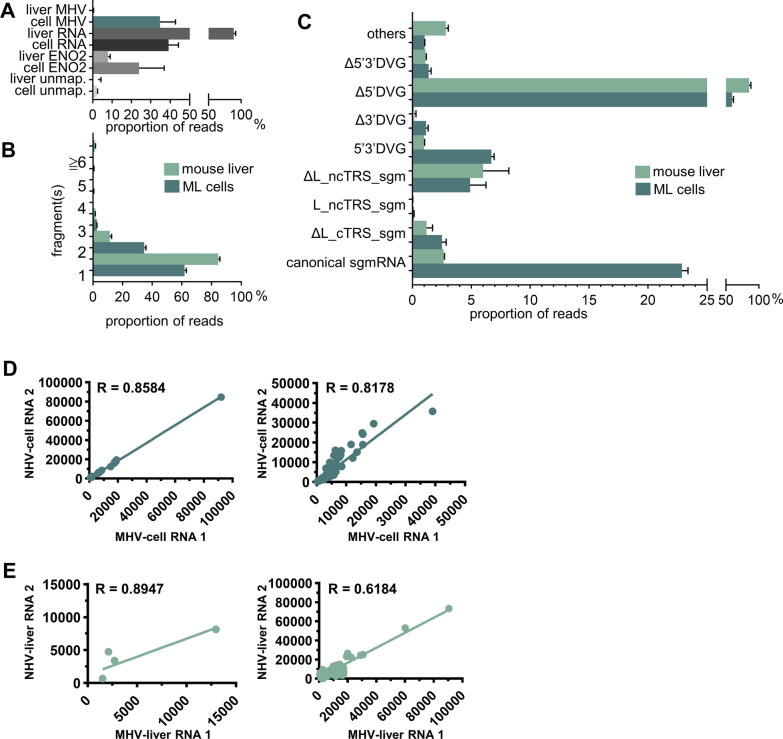
Fig. 5.
Comparison of the biological nature of noncanonical coronavirus transcripts between in vitro and in vivo conditions. A Comparison of the amounts of MHV-A59 transcripts between virus-infected ML cells and mice. cell, ML cells; liver, mouse liver; ENO2, yeast enolase II mRNA; unmap, unmapped. B Comparison of the ratio among transcripts based on the fragment numbers of which the transcripts were composed between ML cells and mice. C Comparison of the ratio of individual MHV-A59 transcripts among the whole transcripts between ML cells and mice. D–E Reproducibility of canonical (left panel) and noncanonical (right panel) transcripts with a read count of ≥ 5 in MHV-A59-infected ML cells (D) and mice (E). The reproducibility was measured as RPKM values and evaluated by Spearman’s correlation coefficient. The data in (A–C) represent the means of two independent experiments. MHV, mouse hepatitis virus-A59; ENO2, Enolase II; unmap, unmapped; DVG, defective viral genome; sgm, subgenomic mRNA; L, leader; c, canonical; nc, noncanonical; ML cells, mouse L cells. ΔL. cTRS. sgm, leader-less sgmRNA derived from canonical TRS; ΔL_cTRS_sgm, leader-less sgmRNA derived from canonical TRS; L_ncTRS_sgm, sgmRNA derived from noncanonical TRS; ΔL_ncTRS_sgm, leader-less sgmRNA derived from noncanonical TRS; 5′3′DVG, DVG with sequence elements from 3′ UTR and 5′ UTR; Δ5′DVG, DVG with a sequence element from 3′ UTR; Δ3′DVG, DVG with a sequence element from 5′ UTR; Δ5′3′DVG, DVG lacking sequence elements from 3′ UTR and 5′ UTR