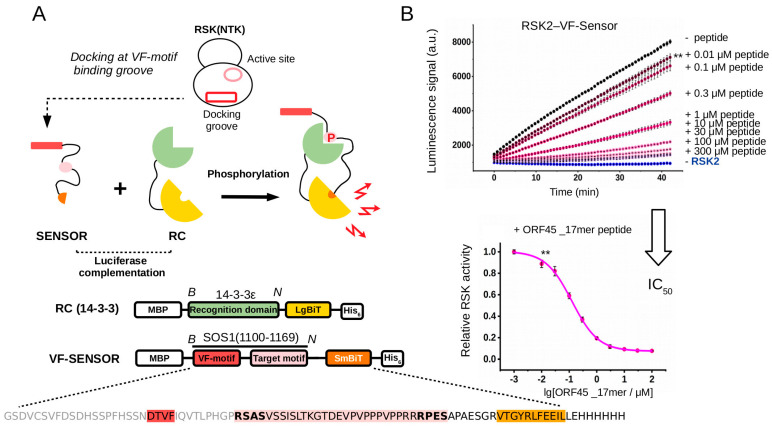
Figure 2.
Principle of the PhALC assay and its validation on RSK-docking-motif-mediated binding. (A) The sensor construct contains the SOS1(1100–1166) region that harbors the VF-motifs and the RxxS/T target sites (VF-Sensor). The latter is bound by the recognition construct (RC) upon phosphorylation. The RC harbors a phosphoserine/-threonine-binding 14-3-3ε domain and the phosphorylation-dependent interaction between the VF-Sensor and RC will facilitate the formation of active luciferase. (The VF-Sensor and the RC are tagged with SmBit and LgBiT fragments of the NanoBiT luciferase, respectively, and also contain N- and C-terminal affinity tags for fast purification; MBP—maltose binding protein, His6—hexahistidine tag). The VF-Sensor and the RC were used in 1 μM and 0.5 μM concentrations, full-length active RSK2 was used in 20 nM concentration and the reaction was started by the addition of ATP. (B) Panels show that the kinetic rate of the luminescence signal correlated with the degree of VF-Sensor phosphorylation by RSK. A VF-motif-containing peptide added in trans competed for binding with the VF-Sensor and thus decreased the rate of the luminescence signal (upper panel). RSK activity was compared to the rate of the reaction containing no peptide (−peptide; the panel shows the raw kinetic luminescence signal; a.u. artificial units; −RSK2: no kinase added). The IC50 value of peptides can be determined when relative RSK activity is plotted as the function of peptide concentration added in trans (lower panel showing the results obtained with ORF45_17mer peptide). Error bars show SD based on three measurements. (B and N denote BamHI and NotI cloning sites with which the functional elements of the sensor and the RC could be modified in the DNA expression plasmids.) Asterisks, here shown only for the lowest concentration of the competitor peptide compared to “−peptide”, indicate results of a two-sided, unpaired t-test (**: p < 0.01).