Figure 1:
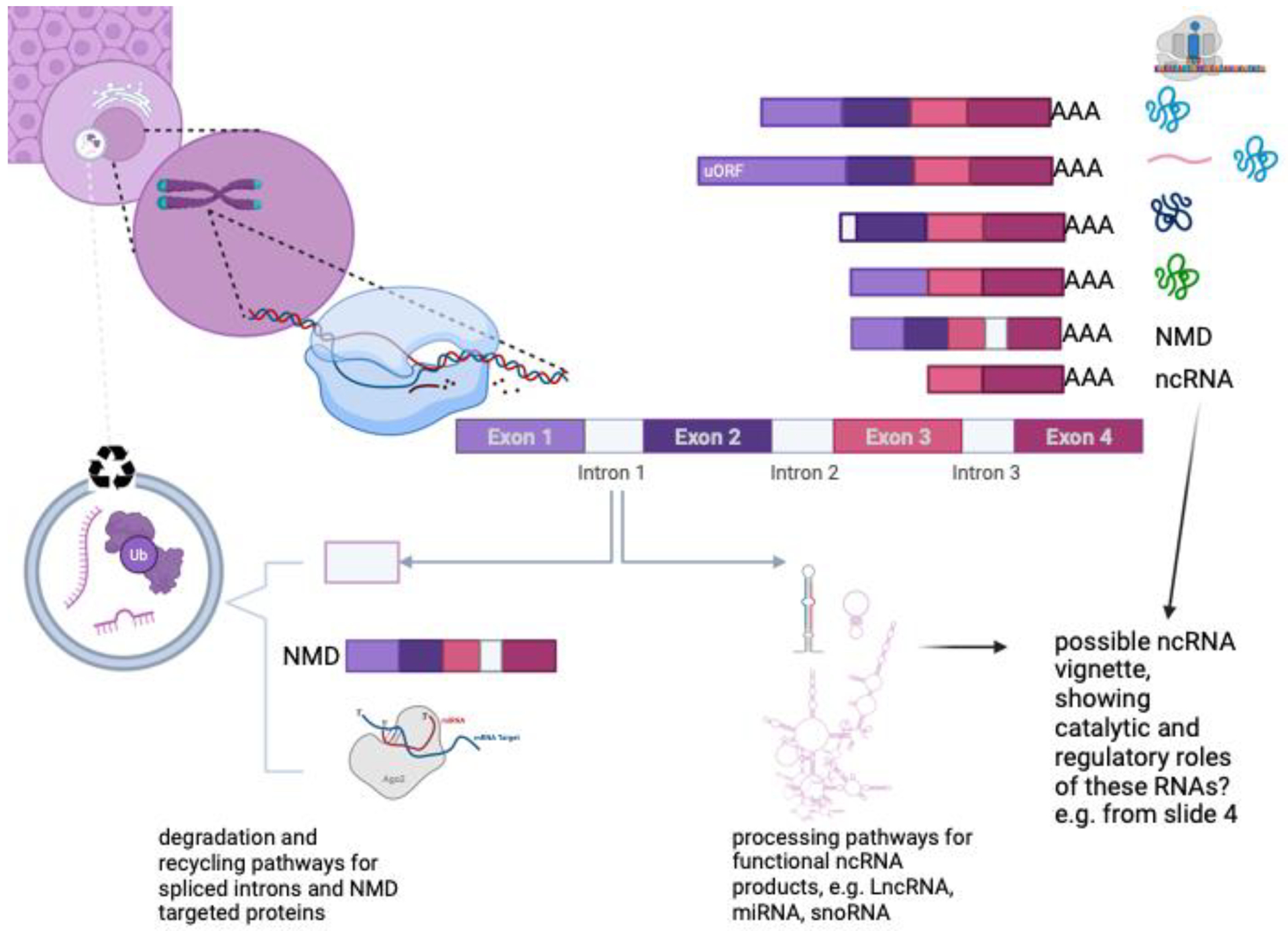
A major challenge for gene annotation is how to capture the diversity of gene products and functions. For example, although the vast majority of protein-coding genes occur on distinct transcripts, a small number of bi-cistronic transcripts encode two distinct open reading frames on the same transcript. Similarly, introns within protein-coding genes may host noncoding RNAs, including miRNAs, snoRNAs or lncRNAs, which may regulate the transcriptional activity of the locus, or may have catalytic roles unrelated to the main protein product. Alternate splicing of transcripts may give rise to proteins that enhance or inhibit each other. Transcripts that are truncated and cannot produce functional proteins are targeted for nonsense-mediated decay (NMD). These products, together with ubiquitinated proteins (Ub) or unwanted intronic material are rapidly recycled by cellular lysozomes. Other seemingly nonproductive transcripts may be repurposed as functional ncRNAs.
