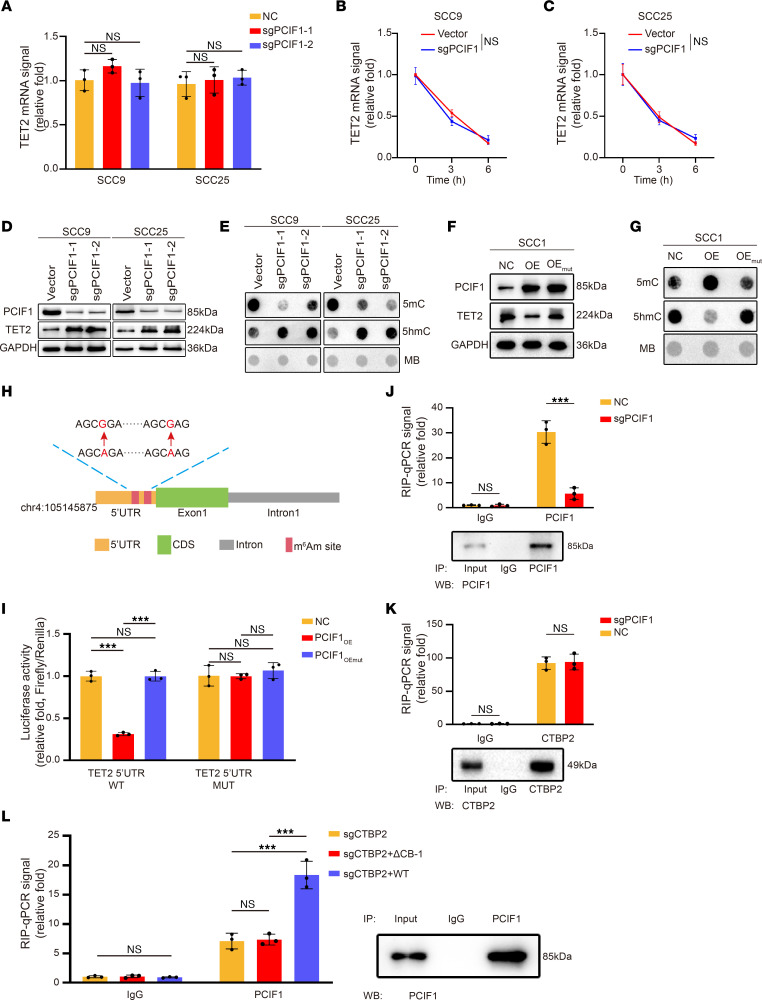
Figure 5. TET2 cap-adjacent m6Am modification suppresses the translation of TET2 mRNA, and PCIF1 requires CTBP2 to bind TET2 mRNA.
(A) Real-time qPCR analysis of TET2 mRNA expression in control and PCIF1-KO cells (n = 3). P > 0.05 by 1-way ANOVA with Tukey’s multiple-comparison test. (B and C) Real-time qPCR analysis of TET2 mRNA levels at the indicated times in control and PCIF1-KO SCC9 (B) and SCC25 (C) cells after actinomycin D treatment (n = 3). P > 0.05 by 2-tailed unpaired Student’s t test. (D and E) TET2 expression (D) and DNA 5mC and 5hmC modification levels (E) were detected in control and PCIF1-KO cells. (F and G) TET2 expression (F) and DNA 5mC and 5hmC modification levels (G) were detected in SCC1 control cells and cells transfected with WT (OE) or mutant PCIF1 plasmid (OEmut). MB, Methylene blue. (H) Schematic diagram of TET2 5′-UTR m6Am site mutations. Red arrows denote A594G and A604G mutation. (I) Luciferase activity of TET2 5′-UTR WT or TET2 5′-UTR m6Am site mutation (5′-UTR MUT) in SCC1 control cells and cells transfected with WT (OE) or mutant PCIF1 plasmid (OEmut) (n = 3). P > 0.05, ***P < 0.001 by 1-way ANOVA with Tukey’s multiple-comparison test. (J and K) RNA immunoprecipitation (RIP)–qPCR analysis of TET2 mRNA retrieved by anti-PCIF1 (J) and anti-CTBP2 (K) antibody in control and PCIF1-KO cells (n = 3). P > 0.05, ***P < 0.001 by 2-tailed unpaired Student’s t test. (L) RIP-qPCR analysis of TET2 mRNA retrieved by anti-PCIF1 antibody in CTBP2-KO cells transfected with vector (sgCTBP2), PCIF1 binding–defective mutant of CTBP2 (ΔCB-1), and WT CTBP2 (n = 3). P > 0.05, ***P < 0.001 by 1-way ANOVA with Tukey’s multiple-comparison test.