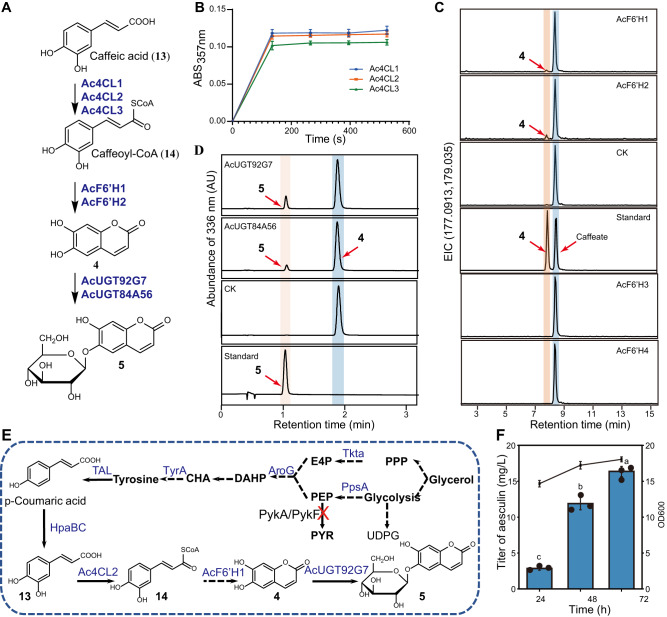
Fig. 4. Functional identification of Ac4CLs, AcF6’Hs and AcUGTs involved in aesculin (5), and de novo biosynthesis of 5.
A Enzymes characterized for the biosynthetic pathway of 5. B Ac4CL enzymes activity assay. Ac4CL1-3 isoforms were assayed for activity against caffeic acid (13) substrate. Enzyme activity was measured by recording the absorbance at 375 nm of caffeoyl-CoA (14) product formation over time. Greater absorbance indicates higher Ac4CL enzyme activity. The data are presented as means values ± s.d. (n = 3 biologically independent samples). C AcF6’H enzymes activity assay. E. coli cells expressing either AcF6’H1 or AcF6’H2 converted substrate 14 into product 4. LC–MS spectrometry detected a peak with the same retention time and mass spectrum as the standard of 4. Standard: 4 and caffeate; CK: E. coli carrying the empty vector. D UPLC analysis for AcUGT92G7 or AcUGT84A56 enzyme activity toward 4; CK: E. coli carrying the empty vector. E The artificial pathway for the biosynthesis of 5 from tyrosine in E. coli. Dashed arrows represent multistep conversion. The red cross line represents the pathway block; genes in blue indicate overexpression. PPP pentose phosphate pathway; UDPG UDP-α-d-glucose; E4P d-erythrose-4-phosphate; PEP phosphoenolpyruvate; PYR pyruvate, DAHP 3-deoxy-arabino-heptulonate-7-phosphate, CHA chorismate, Tkta transketolase, PpsA phosphoenolpyruvate synthase, PykA/pykF pyruvate kinase, AroG phospho-2-dehydro-3-deoxyheptonate aldolase, TyrA chorismate mutase-prephenate dehydrogenase, TAL tyrosine ammonia lyase, HpaBC 4-hydroxyphenylacetic acid 3-hydroxylase, 4CL1 4-coumarate-CoA ligase, F6′H feruloyl-CoA 6′-hydroxylase, AcUGT84A56 and AcUGT92G7 glucosyltransferases. F De novo biosynthesis of 5 by strain XC-6 (BW-1, pCS-TPTA-HpaBC, and pET-648T). The data are presented as means values ± s.d. (n = 3 biologically independent samples). Different letters (a–c) above columns represent significant differences between samples using one-way ANOVA at P < 0.05. Source data are provided as a Source Data file.