Fig. 3.
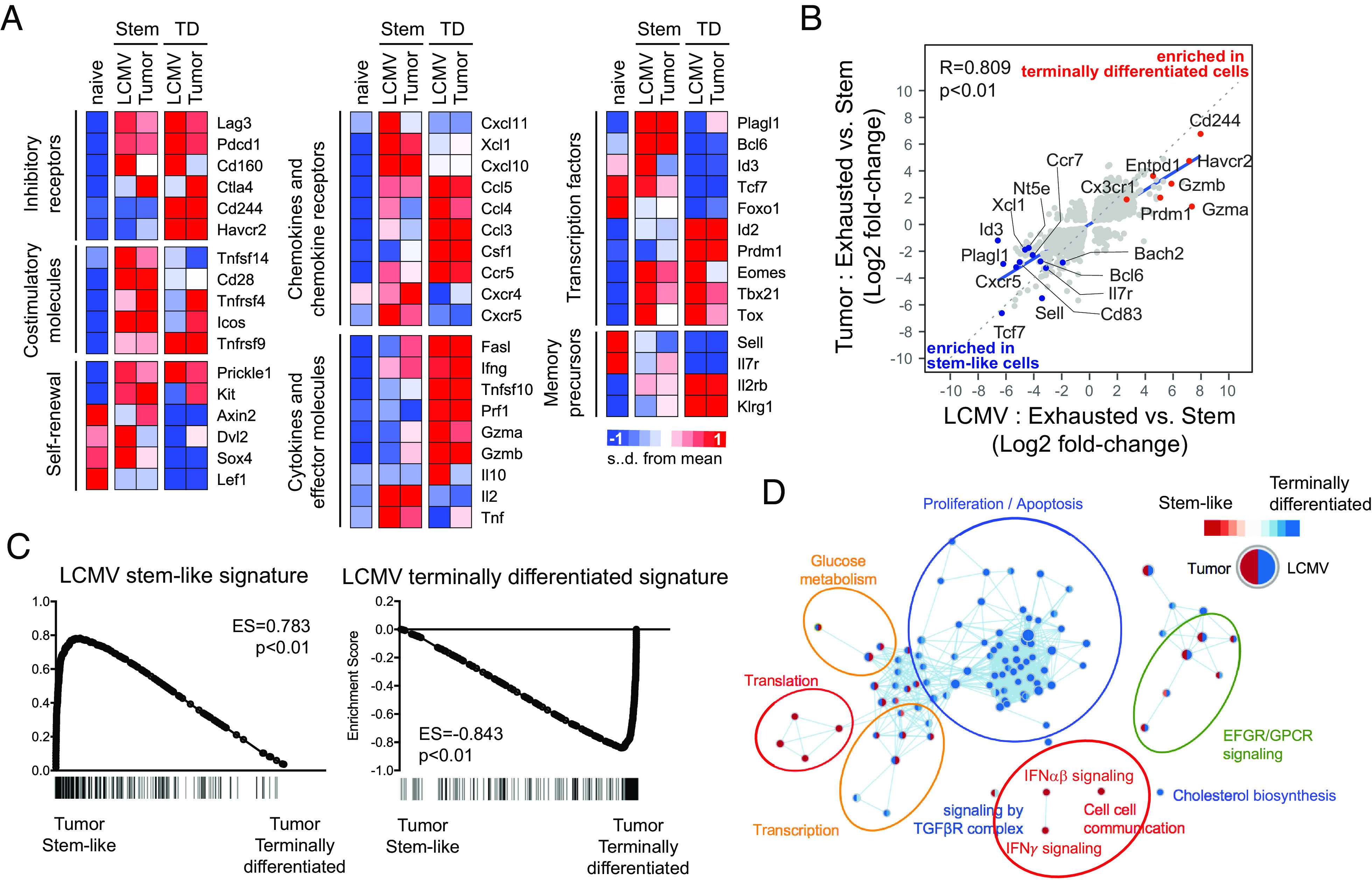
Similarities in transcriptional profiles of CD8 T cell subsets between tumor and chronic LCMV infection. Tim-3−2B4− PD-1+ stem-like and Tim-3+2B4+ PD-1+ terminally differentiated CD8 TILs were isolated from B16F10-GP tumor-bearing mice at day 19 post inoculation. Affymetrix microarray was performed to examine the transcriptome of isolated CD8 TILs, then they were compared with that of two CD8 T cell subsets obtained from mice chronically infected with LCMV (GSE84105) (A) Heat map displaying the relative expression of genes in naive (CD44loCD127+CD62L+) CD8 T cells from uninfected mice, two CD8 TIL subsets from tumor-bearing mice and two CD8 T cell subsets from chronically infected mice. (B) A scatter plot showing differentially expressed genes with twofold cutoff (in addition to the adjusted P-value < 0.05 cutoff) between two CD8 T cell subsets in chronically infected mice (X axis) and tumor-bearing mice (Y axis). (C) GSEA for identifying specific gene signatures of two murine CD8 TIL subsets compared to gene signatures of LCMV stem-like and exhausted CD8 T cells. (D) Cytoscape network analysis for identifying enriched Reactome pathways in each CD8 T cell subset.
