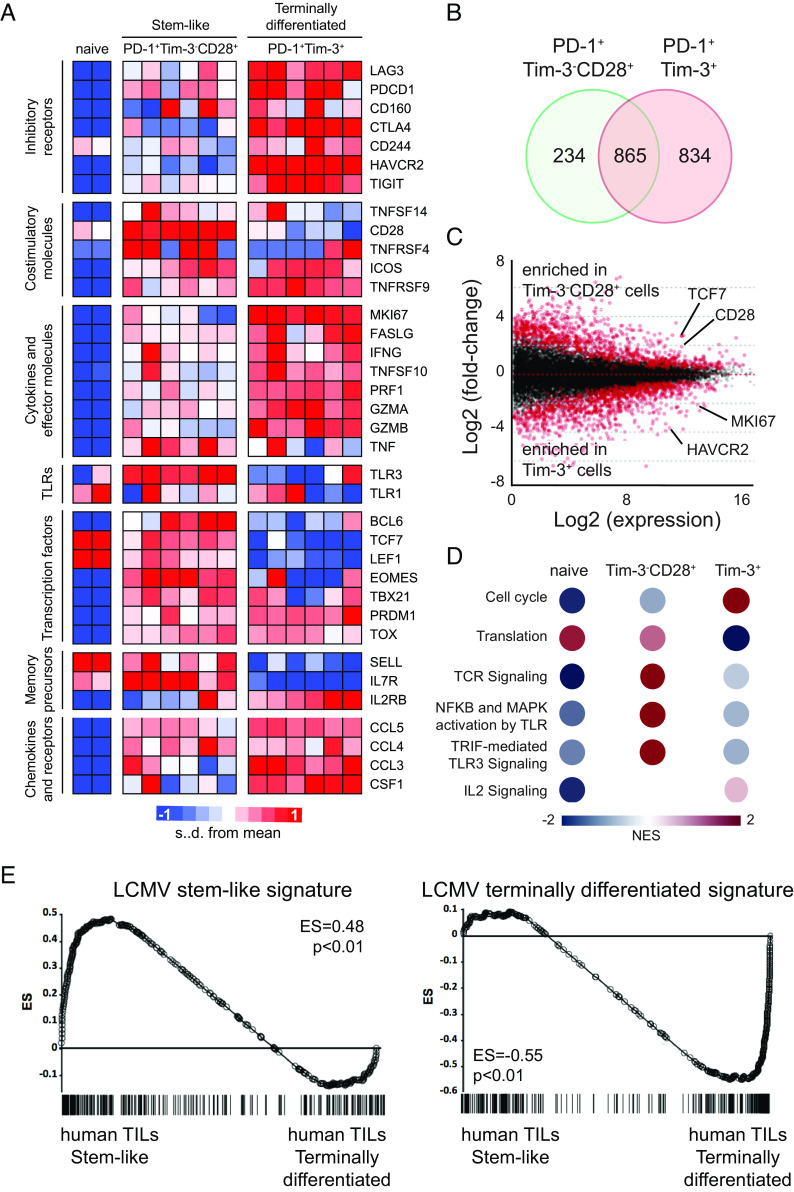
Fig. 7.
Transcriptional profiles of two CD8 TIL subsets isolated from NSCLC patients. Tim-3−CD28+ PD-1+ stem-like and Tim-3+ PD-1+ terminally differentiated CD8 TILs were isolated from NSCLC patients. RNA-seq was performed to examine the transcriptome of isolated CD8 TILs. (A) Heat map displaying the relative expression of genes in naive (CCR7+CD45RA+CD58−CD95−) CD8 T cells from a healthy subject and a NSCLC patient and two CD8 TIL subsets from six NSCLC patients. (B) DEGs in the stem-like and terminally differentiated CD8 T cells compared to naive CD8 T cells. Venn diagram illustrates the overlap in stem-like and exhausted cells among DEGs. (C) Scatter plot of log2 fold-change between stem-like and terminally differentiated CD8 T cells versus mean average read counts. Red indicates genes that were significantly different (adjusted P-value < 0.01). (D) Reactome pathways which were enriched in each CD8 T cell subset compared to other two subsets. NES, normalized enriched score. (E) GSEA for identifying specific gene signatures of two human CD8 TIL subsets compared to gene signatures of LCMV stem-like and terminally differentiated CD8 T cells.