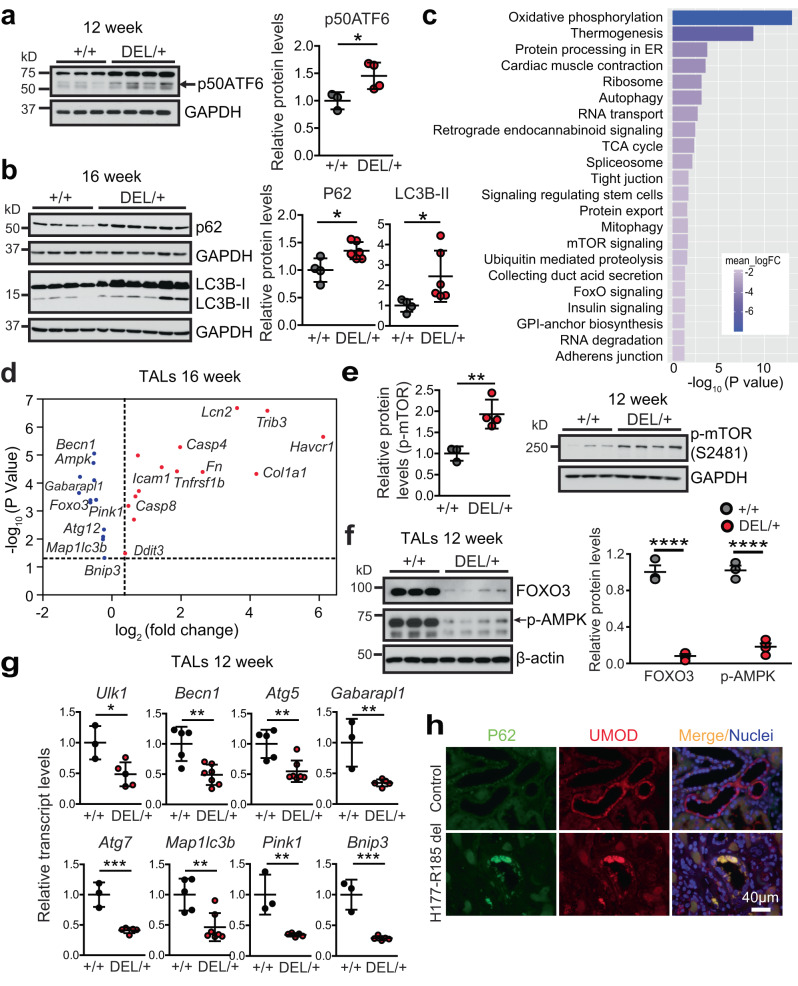
Fig. 2. Impaired autophagy in the mutant TALs in ADTKD-UMOD.
a Whole-kidney lysates from Umod +/+ (n = 3) and Umod DEL/+ (n = 4) mice at 12 weeks were analyzed by WB for p50ATF6 (arrow) with densitometry analysis. Two-tailed t-test, Mean ± SD. *p = 0.0376. b WBs of whole-kidney lysates (WT, n = 4; Umod DEL/+, n = 6) at 16 weeks to detect the autophagy mediators p62 and LC3B with densitometry analysis. Two-tailed t-test, Mean ± SD. *p = 0.016 (for p62), 0.049 (LC3B-II). c, d RNA-seq analysis of mutant and WT TALs at 16 weeks. n = 5/group. Benjamini-Hochberg adjusted p < 0.05. c KEGG pathway analysis of the log2 fold-changes in TAL cells showed the top 22 downregulated pathways. GAGE analysis. d RNA-seq analysis of genes for autophagy, ER stress response, inflammation, apoptosis, kidney injury and fibrosis in the mutant TALs compared with WT TALs. Limma analysis for differential gene expression. e WB from Umod +/+ (n = 3) and Umod DEL/+ (n = 4) kidneys for levels of p-mTOR (S2481) at 12 weeks with densitometry analysis. Two-tailed t-test, Mean ± SD. **p = 0.0081. f WBs from Umod +/+ (n = 3) and Umod DEL/+ (n = 4) TALs for levels of FOXO3 and p-AMPK (arrow) at 12 weeks with densitometry analysis. Two-tailed t-test, Mean ± SD. ****p < 0.0001. g Transcript analysis of a panel of autophagy and mitophagy-related genes, normalized to 18 s, from TALs at 12 weeks. WT (n = 3) and Umod DEL/+ (n = 5) for Ulk1, Gabarapl1, Atg7, Pink1 and Bnip3; WT (n = 5) and Umod DEL/+ (n = 7) for Becn1, Atg5 and Map1lc3b. Two-tailed t-test, Mean ± SD. p = 0.02 (for Ulk1), 0.0027 (Becn1), 0.0034 (Atg5), 0.0079 (Gabarapl1), 0.0006 (Atg7), 0.0039 (Map1lc3b), 0.0032 (Pink1), 0.0005 (Bnip3). h Representative IF images of human kidney biopsies obtained from a patient with p.H177-R185 del and from a normal kidney, stained for p62 (green) and UMOD (red) with nuclei counterstain (blue) on paraffin kidney sections. Scale bar, 40 µm. Source data are provided as a Source Data file.