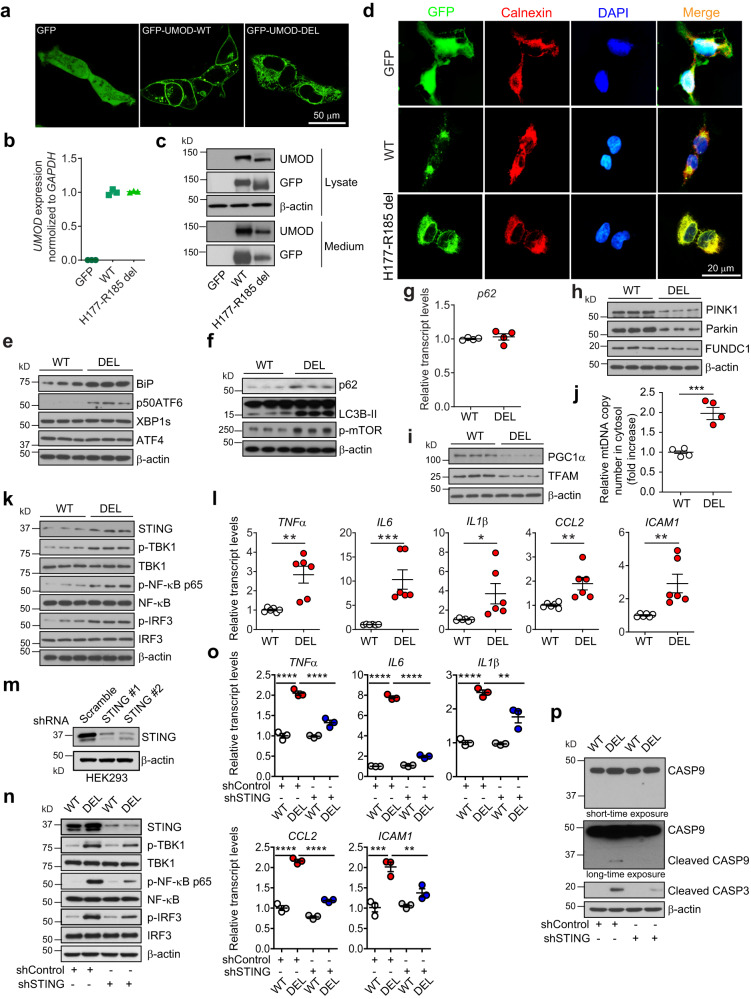
Fig. 5. Cytosolic leak of mtDNA activates STING signaling by utilizing stable HEK 293 cell line expressing N-terminal GFP-tagged WT or H177-R185 del UMOD.
a Confocal live imaging showing GFP signal in HEK 293 cells expressing GFP, GFP-tagged WT or mutant H177-R185 del uromodulin. b UMOD transcript expression assessed by RT-qPCR. Mean ± SD of fold changes of three independent samples from each clone. c Cell lysates and media from the indicated cells were analyzed by WBs for expression of GFP-UMOD by using anti-UMOD or anti-GFP antibody. d IF analysis of GFP and calnexin with nuclei counterstain (DAPI). e, f, h, i, k Cell lysates of WT and DEL cells were analyzed by WBs for the indicated proteins. n = 3/group. g Quantitative PCR analysis for mRNA levels of p62, normalized to GAPDH. n = 4/group. Mean ± SD. j Cytosolic translocation of mtDNA in WT and DEL cells was quantified by q-PCR. The copy number of mtDNA encoding cytochrome c oxidase I was normalized to nuclear DNA encoding 18 S ribosomal RNA. n = 4/group. Two-tailed t-test, Mean ± SD of fold changes. ***p = 0.0008. l Quantitative PCR of proinflammatory genes downstream of STING/NF-κB signaling, normalized to GAPDH. n = 6/group. Two-tailed t-test, Mean ± SD. p = 0.0019 (for TNFα), 0.0009 (IL6), 0.0287 (IL1β), 0.0069 (CCL2), 0.0068 (ICAM1). m WB analysis showed knockdown efficacy of STING expression from shSTING1 and shSTING2 vs. a scrambled shRNA control in HEK 293 cells. n Cell lysates of WT and DEL cells, treated with shControl or shSTING for 48 h, were analyzed by WBs for the indicated proteins. o Quantitative PCR of proinflammatory genes, normalized to GAPDH, in cells treated with shControl or shSTING for 48 h. n = 3/group. One-way ANOVA with Tukey’s multiple comparisons, Mean ± SD. TNFα, IL6, CCL2: ****p < 0.0001; IL1β: ****p < 0.0001, **p = 0.0044; ICAM1: ***p = 0.0003, **p = 0.0062. p Cell lysates of WT and DEL cells, treated with shControl or shSTING for 48 h, were analyzed by WBs for cleaved caspases 9 and 3. Source data are provided as a Source Data file.