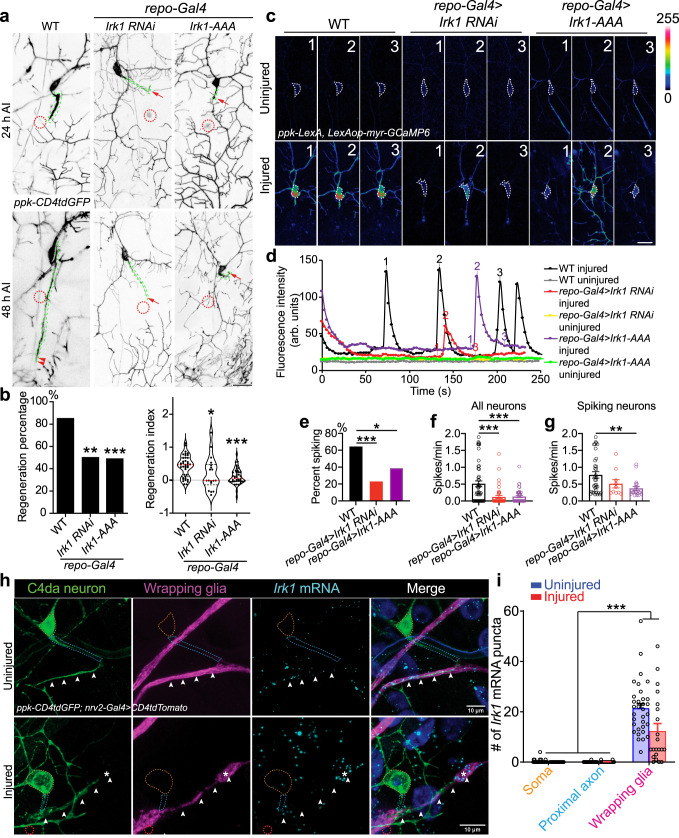
Fig. 5. Irk1 is expressed in glial cells and is required for axon regeneration and axotomy-induced Ca2+ transients.
a, b LoF of glial Irk1 by RNAi or Irk1-AAA OE reduces C4da neuron axon regeneration. Arrow marks axon stalling while arrowheads show the regrowing axon tips. N = 40, 18, 41 neurons, P = 0.0087, 0.0008 (left). c–g Glial Irk1 is required for axotomy-induced Ca2+ transients in C4da neurons. (c) C4da neurons show Ca2+ spikes after axonal injury in WT. LoF of Irk1 by glial RNAi or Irk1-AAA reduces Ca2+ spikes. Images from three timepoints are presented. The dashed white circle marks the cell body. No obvious difference is observed in the uninjured condition. d Plots of the mean fluorescence intensity over time in the cell body. e Quantification of the percent of neurons showing Ca2+ spikes. N = 48, 43, 41 neurons, P = 0.0001, 0.0200. f, g Quantification of the number of Ca2+ spikes per minute in all neurons (f, N = 48, 43, 41 neurons, P = < 0.0001, <0.0001) or spiking neurons (g, N = 35, 10, 20 neurons, P = 0.1203, 0.0062), average values and standard error of the means are shown. h, i Irk1 mRNA is highly expressed in wrapping glia. Irk1 expression is analyzed around the C4da neuron soma (outlined in dotted orange circle), proximal axon (dotted blue circle) and axon stretch (white arrowheads) covered by wrapping glia (nrv2-Gal4). With or without axon injury, Irk1 transcripts are enriched in the wrapping glia along the axon. Axons are observed to regenerate along/towards wrapping glia with high Irk1 expression, marked by asterisks. N = 21, 33 neurons, P = < 0.0001. average values and standard error of the means are shown. *P < 0.05, **P < 0.01, ***P < 0.001, Fisher’s exact test, two-sided (b left and e), one-way ANOVA followed by Holm–Sidak’s test (b right, f, g) or Tukey’s test (i). Scale bar = 20 µm (a, c) and 10 µm (h). Source data are provided as a Source data file.