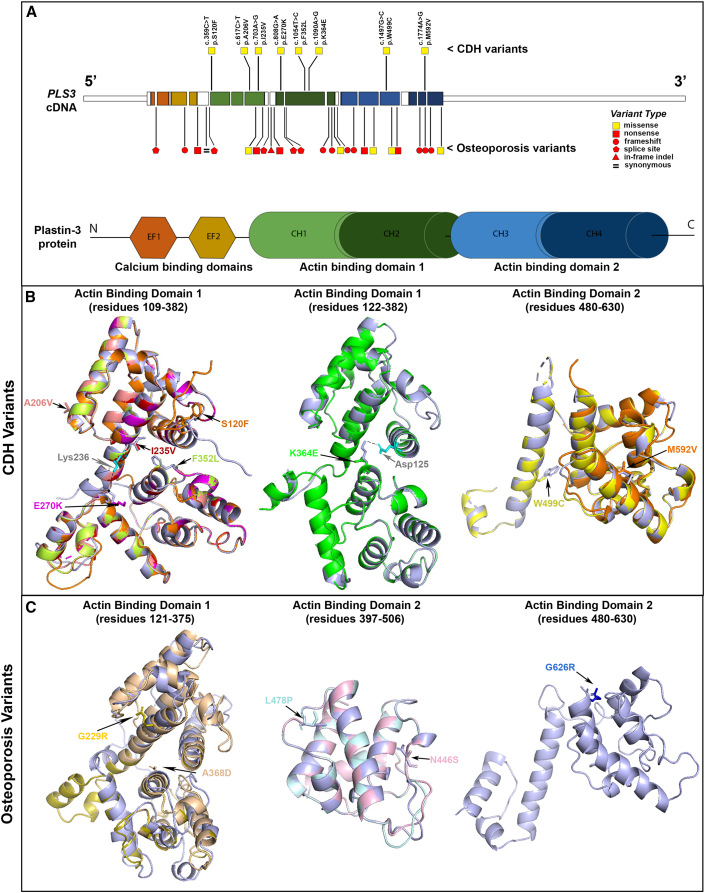
Figure 3.
Structural modeling of human PLS3 variants
(A) Locations and types of PLS3 variants in individuals with CDH and X-linked osteoporosis. The cDNA structure for the canonical PLS3 transcript ENST00000420625.2 or GenBank: NM_005032 is shown at the top, and the color-coded exons depicting the corresponding domains of the Plastin-3 protein are shown at the bottom. The CDH variants are depicted above the cDNA, and the osteoporosis variants16 are depicted below the cDNA. Yellow squares indicate missense variants, and red shapes indicate LoF variants (square, nonsense; circle, frameshift; pentagon, essential splice site; triangle: in-frame indel). The Plastin-3 protein image shows the following domain types: EF, EF-hand domain; CH, Calponin-homology domain.
(B) Predicted PLS3 actin-binding domain protein structures for CDH-associated variants. In these images, the predicted structures of each of the eight variant Plastin-3 proteins (multiple colors) are overlaid on the structure of the native (wild-type) Plastin-3 (lavender). The amino acid alterations are written in the same color as the corresponding protein structure. The overlaid structures demonstrate that none of the CDH variants are predicted to cause a major alteration of protein structure. The dotted lines indicate salt bridges between Lys236 and Glu270 (left) and Asp125 and Lys364 (middle) of the wild-type protein.
(C) Predicted PLS3 actin-binding domain protein structures for osteoporosis-associated missense variants. The predicted structures of five PLS3 variants (yellow, peach, pale blue, pale pink, and blue) are overlaid on the structure of the native protein (lavender), showing that most osteoporosis variants are predicted to cause a major change in protein structure.