Experimental design of NK cell killing assays.
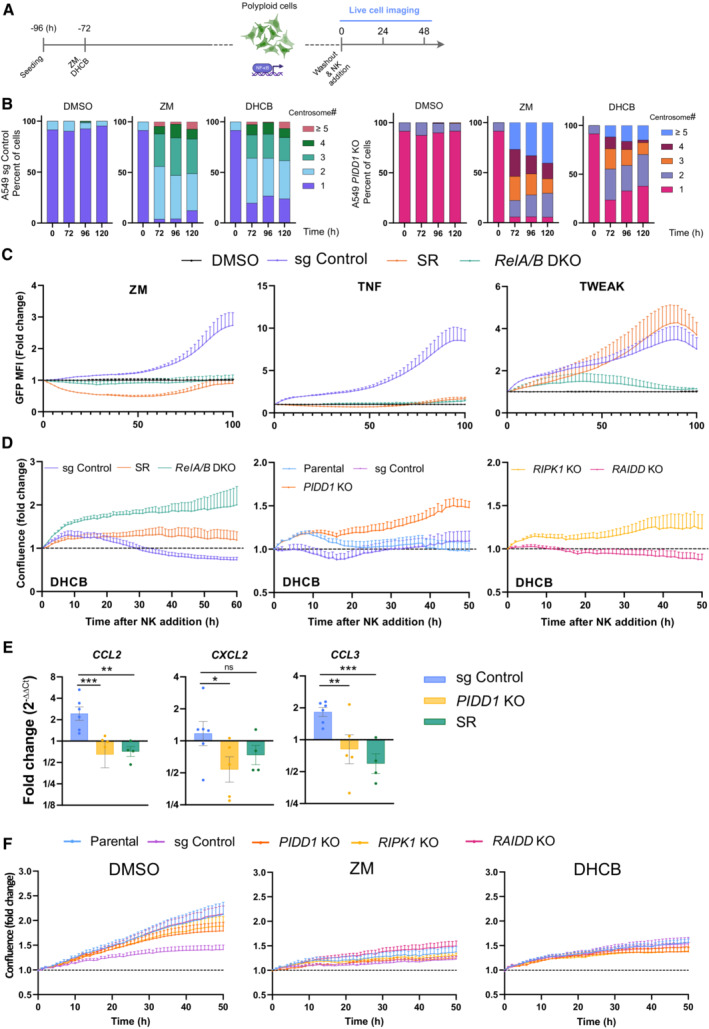
Evaluation of centriole counts based on IF analysis and γ‐tubulin staining. More than 50 cells were evaluated per condition in two independent experiments. Data are presented as means values.
Evaluation of reporter activity in A549 cells of the indicated genotypes proficient or deficient in NF‐κB activation, due to loss of RELA/B or overexpression of the IkBα super‐repressor (SR) after treatment with ZM, the canonical NF‐κB pathway agonist, TNF or the non‐canonical NF‐κB pathway agonist TWEAK. n = 3 independent experiments; mean ± SEM.
Confluence curves from A549EGFP derivatives of the indicated genotypes, pretreated with DHCB (4 μM) to induce cytokinesis defects and co‐cultured with NK‐92 cells. n ≥ 3 independent experiments; mean ± SEM.
Conditioned medium from ZM pretreated A549 cells of the indicated genotypes was applied to human monocyte–derived macrophages for 24 h followed by RNA isolation and qRT‐qPCR analysis of macrophage activation markers. Expression values are plotted as the fold‐change relative to macrophages stimulated with conditioned medium from untreated A549 cells. HPRT1 expression was used for normalization. Bars represent mean ± SEM of n ≥ 4 independent experiments. The statistical significance was determined using two‐way ANOVA; P ≤ 0. 1 (*), P ≤ 0.01 (**), P ≤ 0.001 (***).
Proliferation curves of different A549EGFP derivatives, pretreated with ZM (2 μM) or DHCB (4 μM), for 72 h in the absence of NK cells. Time zero is defined as the moment of the drug wash out. Live‐cell imaging of target cells without NK cells was performed in parallel to killing experiments using the same conditions as described in the methods section. n = 5 independent experiments; mean ± SEM.