FIGURE 3.
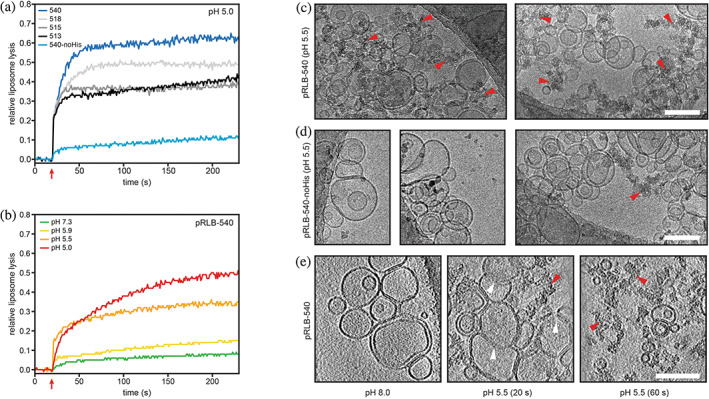
Liposome disruption by pH‐responsive lytic bundles (pRLBs). (a and b) Proteins were incubated with synthetic liposomes encapsulating sulforhodamine B (SRB) at self‐quenching concentrations; liposome disruption was investigated by measurement of SRB fluorescence. Red arrows indicate acidification to the respective pH. (a) All four designs tested were able to lyse liposomes at pH 5 to different extent and plateaued after 60 s. Mutation of the histidine residue in the pH‐responsive hydrogen bond network to asparagine impairs lytic activity. (b) Liposome lysis of pRLB‐540 is tightly controlled by pH. Cryo‐electron microscopy was used to visualize liposomes mixed with pRLB‐540 (c) and pRLB‐540‐noHis (d). Representative images of the respective condition are shown. The mixture was brought to pH 5.5, and plunge‐frozen after a 60 s incubation, and imaged by conventional cryo transmission electron cryomicroscopy. Cryo‐electron tomography was used to image pRLB‐540 mixed with liposomes at pH 8.0, and after 20 and 60 s incubation at pH 5.5 (e). Images shown in panel (e) are averages of 17 Å thick slices in the Z‐plane. White arrowheads indicate liposome membranes in an intermediate state of disruption. Red arrowheads indicate clusters of electron dense aggregate material. All scale bars 100 nm.
