Figure 5. Nasal airway DEmiRNAs pathways and risk of developing asthma.
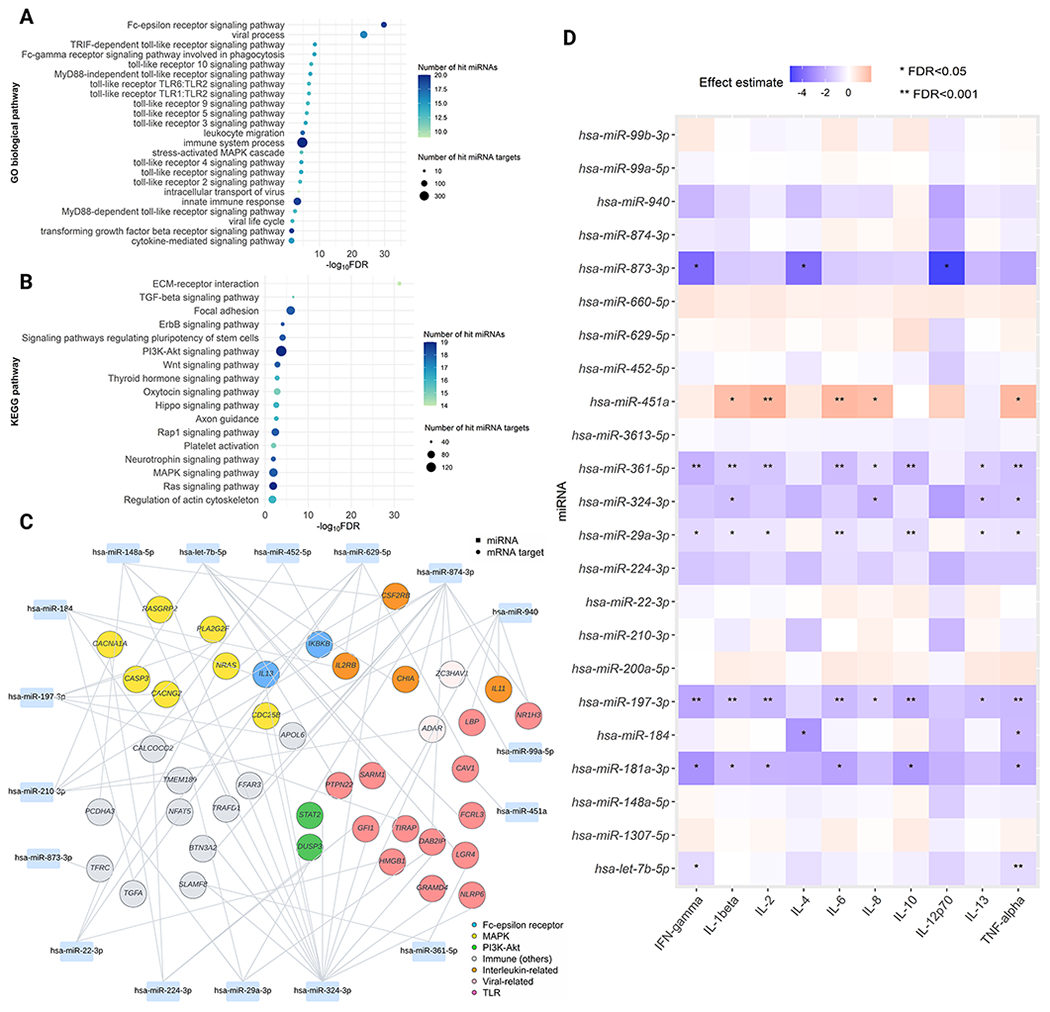
A. Functional enrichment analysis using GO biological process gene sets We identified 108 differentially-enriched pathways (FDR<0.05) associated with the asthma risk. Of these, we selected immune-related pathways to visualize the plot. B. Functional enrichment analysis using KEGG gene sets We identified 41 differentially-enriched pathways (FDR<0.05) associated with the asthma risk. Of these, we selected non cancer-related pathways to visualize the plot. C. DEmiRNA-mRNA target network A rectangle denotes miRNA, a circle denotes mRNA target. mRNA targets that are mapped within major immune-related pathways are highlighted in various colors. In this network, we only included mRNA targets that are negatively correlated (Spearman correlation coefficient<−0.1) with DEmiRNAs and have immune-related functions based on prior knowledge. D. Association of the 23 DEmiRNAs with the ten nasal cytokines. The blue-to-red gradient in the heatmap denotes the magnitude and direction of the associations. Abbreviations: FDR, false discovery rate; IFN, Interferon; IL, interleukin; MAPK, Mitogen-activated protein kinase; PI3K-Akt, phosphatidylinositol 3‑kinase-protein kinase B; TLR, Toll-like receptor; TNF, Tumor necrosis factor.
