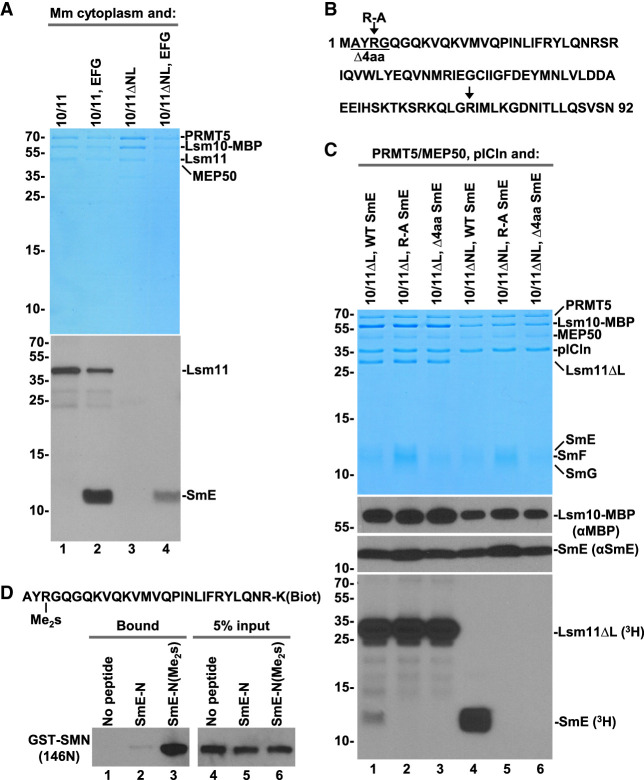
FIGURE 5.
In vitro methylation of SmE by endogenous methylosome and identification of the SmE methylation site. (A) Recombinant Lsm10/11 (lanes 1,2) and Lsm10/11ΔNL (lanes 3,4) heterodimers were incubated either in the absence or in the presence of the SmE/F/G heterotrimer with a mouse cytoplasmic extract and purified together with bound endogenous methylosome on amylose beads via MBP attached to Lsm10. The immobilized proteins were tested directly on the beads in a buffer containing 3H SAM for the ability to methylate Lsm11 and SmE. Following overnight methylation, proteins in each sample were stained with Coomassie Blue (top panel) and their methylation status analyzed by fluorography (bottom panel). (B) Sequence of human SmE (amino acids 1–92) and mutations made near the amino terminus. The arrows indicate each of the two arginines in SmE neighboring a glycine. (C) Recombinant methylosome complex consisting of PRMT5, MEP50, and pICln was incubated with Lsm10/11ΔNL heterodimer in the presence of 3H SAM and SmE/F/G heterotrimer containing WT, R-A or Δ4aa variants of SmE. Proteins used in the assay were resolved by SDS-PAGE and visualized by staining with Coomassie Blue (top panel) and their methylation status analyzed by fluorography (bottom panel). Lsm10-MBP and SmE were additionally detected by western blotting using αMBP and αSmE antibodies, respectively (middle panels). (D) Two carboxy-terminally biotinylated peptides encompassing the first 27 amino acids of SmE and either lacking or containing symmetric dimethyl group at Arg4 were incubated with a GST-tagged amino-terminal region of SMN (amino acids 1–146) containing the Tudor domain. Proteins immobilized on streptavidin beads were resolved by SDS-PAGE and probed using anti-GST antibody. Lane 1 represents the background observed in the absence of any biotinylated peptide. The input (5%) for all binding reactions is shown in lanes 4–6.