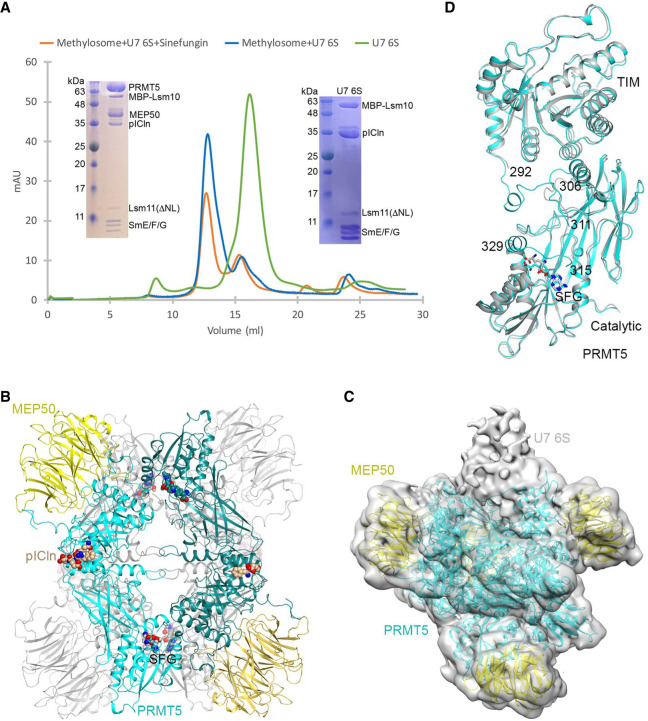
FIGURE 6.
Structural studies of the human methylosome–U7 6S complex. (A) Gel filtration profiles of Lsm10/11 bound to SmE/F/G and pICln (U7 6S complex, green), methylosome bound to U7 6S (blue), and methylosome bound to U7 6S in the presence of sinefungin (SFG, orange). Inserts are SDS gels of the purified U7 6S sample (right) and methylosome–U7 6S sample (left). Lsm10 is amino-terminally fused with MBP, and the amino-terminal region and internal loop of Lsm11 are deleted (Lsm11ΔNL). (B) Schematic drawing of the human methylosome–U7 6S complex. Only the PBM of pICln is included in the atomic model, and the rest of the U7 6S is flexible. (C) Cryo-EM density for the methylosome–U7 6S complex, low pass filtered to 7 Å resolution. The atomic model for the methylosome is shown. The extra EM density that could correspond to U7 6S is labeled. (D) Overlay of two PRMT5 molecules with different conformations in the methylosome–U7 6S complex. The PRMT5 molecule in contact with the putative U7 6S EM density is shown in cyan and the other molecule in gray. The disordered segments, 292–306 and 311–315 or 311–329, are labeled. Panel D was produced with Chimera (Goddard et al. 2007).