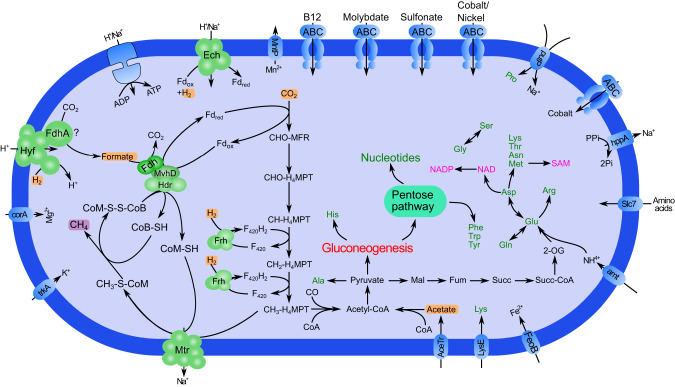
Fig. 6. Metabolic map of Ca. Methanoregula pelomyxae reconstructed from the draft genome.
Amino acids are shown in green; vitamins and cofactors are shown in pink. Non-standard abbreviations: 2OG 2-oxoglutarate, CH3-S-CoM methyl coenzyme M, CHO-MFR formylmethanofuran, CO carbon monoxide, H4-MPT Tetrahydromethanopterin, CH-H4MPT 5,10-Methenyltetrahydromethanopterin, CH2-H4MPT 5,10-Methylenetetrahydromethanopterin, CH3-H4MPT 5-Methyltetrahydromethanopterin, CoA coenzyme A, CoB-SH Coenzyme B, CoM-SH Coenzyme M, CoM-S-S-CoB Coenzyme M 7-mercaptoheptanoylthreonine-phosphate heterodisulfide, Fum fumarate, Mal malate, MFR methanofuran, SAM S-adenosylmethionine, Succ succinate, Succ-CoA succinate coenzyme A. Putative metabolic end products are highlighted with a purple background. Metabolites that are putatively used from the host and/or environment are highlighted with an orange background.