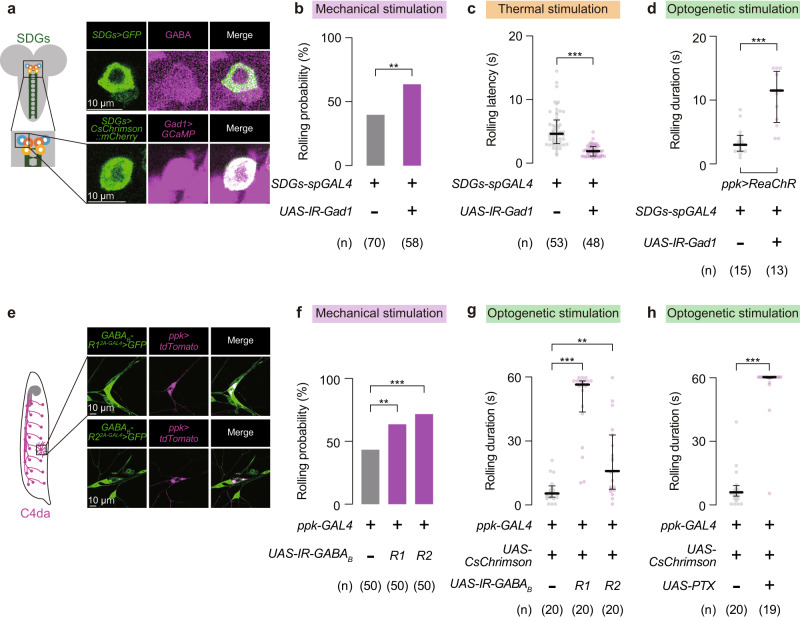
Fig. 3. SDGs negatively control rolling behavior through GABAergic inputs to C4da neurons.
a Top, immunohistochemistry of GABA (magenta) at the cell body of SDGs labeled with GFP (green). Bottom, dual detection of GCaMP and CsChrimson::mCherry signals driven by Gad1-LexA (magenta) and SDGs-spGAL4 (green), respectively. Similar results were obtained across 5 independent samples per genotype. b–d Behavioral phenotypes caused by Gad1 knockdown in SDGs. C4da neurons were stimulated by three distinct approaches. For mechanical stimulation (b), rolling probability is shown in the bar graph. Note that the background genotypes of parental flies used in b were different from other experiments; see Supplementary Table 1 for full genotypes. In this and following panels, ‘n’ indicates the number of biologically independent animals used for each group, and the thick line and the thin error bar represent the median and interquartile range, respectively. **p < 0.005 (Fisher’s exact test). For thermal (c) and optogenetic (d) stimulation, latency and duration are each indicated. ***p < 0.0005 (Mann–Whitney U-test). e Colocalization of GFP driven by either GABAB-R12A-GAL4 or GABAB-R22A-GAL4 (green) with ppk-CD4-tdTomato (magenta) in C4da cell bodies. Similar results were obtained across 4 independent samples per genotype. f, g Nociceptive responses in larvae expressing RNAi constructs targeting each GABAB-R gene in C4da neurons. Rolling probabilities after the mechanical stimulation are shown in the bar graph (f). ***p < 0.0005, **p < 0.005 (Fisher’s exact test with Bonferroni correction). For optogenetic experiments (g), rolling durations within the 60-s stimulation period are indicated. ***p < 0.0005, **p < 0.005 (Mann–Whitney U-test with Bonferroni correction). h Effect of PTX expression on larval rolling induced by optogenetic C4da activation. ***p < 0.0005 (Mann–Whitney U-test). See Supplementary Table 1 for full genotypes. Source data are provided as a Source Data file.