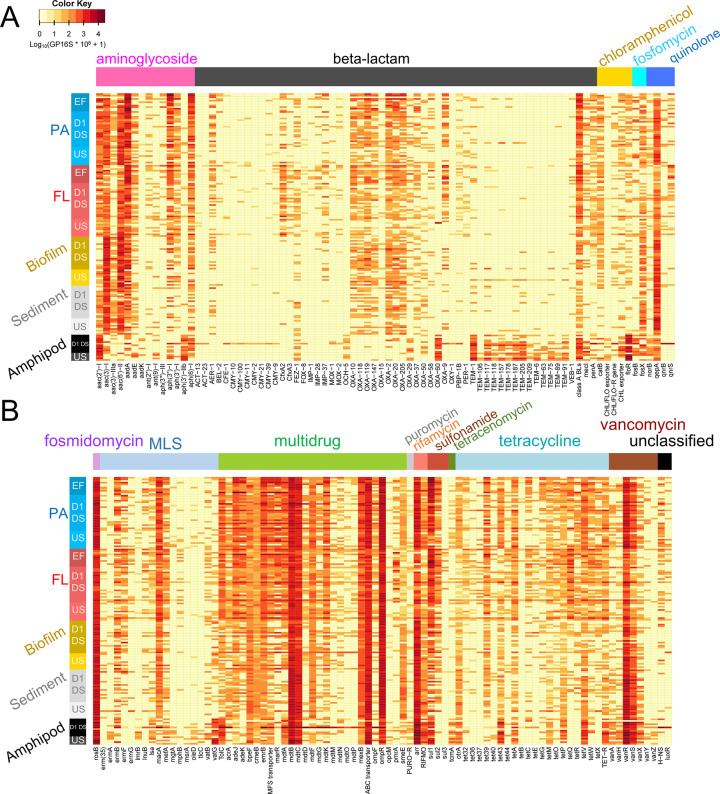
Fig. 3. Heatmaps displaying relative abundances of selected ARGs selected based on their explanatory power in ordination (see Fig. 2) in the metagenomes of different river habitats.
A ARGs conferring aminoglycoside, beta-lactam, chloramphenicol, fosfomycin and fluoroquinolone resistance. B ARGs conferring macrolide-lincosamide-streptogramin (MLS), multidrug, sulfonamide, tetracycline and other resistances. Relative abundance was log-transformed (i.e., Log10(GP16S × 106 + 1)) for the plot. Each row indicates a sample (arranged by sample type). PA denotes particle-associated (PA, > 5.0 μm) wastewater (EF) or river water biomass from different locations (D1, DS, US) and FL denotes free-living (FL, 0.2–5.0 μm) wastewater or river water biomass. BLs, CHL, FLO, MFS, PURO, RIFMO, and TET stand for beta-lactamase, chloramphenicol, florfenicol, major facilitator superfamily, puromycin, rifampin monooxygenase, and tetracycline, respectively. ‘-R’ indicates ‘-resistance gene’.