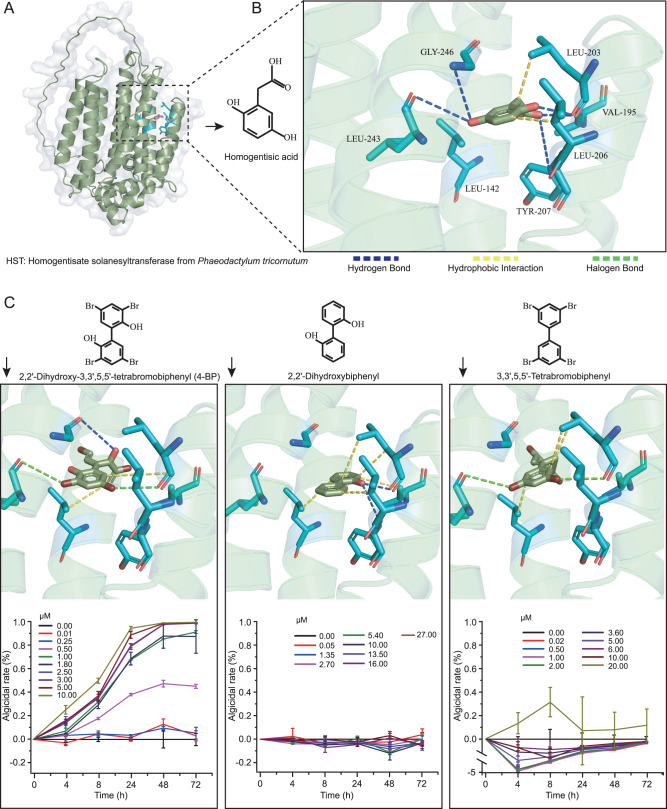
Fig. 6. Structure prediction of homogentisate solanesyltransferase (HST) from Phaeodactylum tricornutum CCMP2561 and its substrate docking.
A The three-dimensional structure of the HST sequence was constructed using ColabFold; (B) Docking studies (with AutoDock) showed the binding of homogentisic acid to HST. Amino acids at the HST active site are shown such that: LEU leucine, GLY glycine, VAL valine, and TYR tyrosine. Homogentisic acid docked inside the active site and linked to these amino acids with different interactions; (C) The binding of three structurally similar molecules (i.e., 4-BP, 2,2’-dihydroxybiphenyl and 3,3’,5,5’-tetrabromobiphenyl) to the HST are shown along with their algicidal effect on Phaeodactylum tricornutum CCMP2561, when tested at different concentrations.