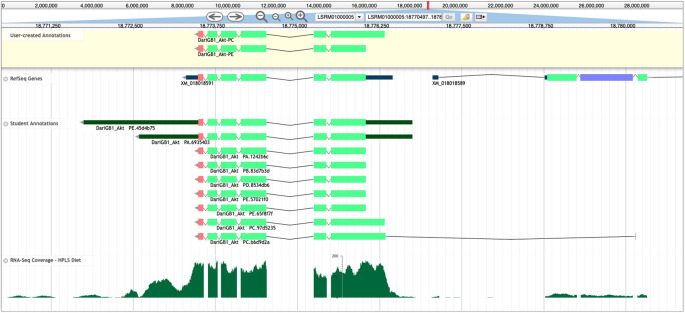
Figure 6. Apollo Screenshot for the Akt gene model in D. arizonae.
Final gene models for Akt in D. arizonae (User-created Annotations track, yellow background), along with the NCBI RefSeq gene model (RefSeq Genes track), submitted student models (Student Annotations track), and RNA-Seq data aligning to the region (RNA-Seq Coverage – HPLS Diet track, histograms). Despite RefSeq predicting only one isoform (XM_018018591) for this gene, the final model contains two unique protein-coding isoforms (Akt-PC and Akt-PE), which were annotated using multiple lines of evidence. The Akt-PC isoform has a larger coding region in the reconciled gene model that is missed by the RefSeq gene predictions. In the Student Annotations track, the top two models included annotations of the 5’ and 3’ untranslated regions (UTRs, thinner dark green rectangles), and the bottom student model (DariGB1_Akt-PC.b6d9d2a) incorporated a coding exon that was likely not part of Akt.