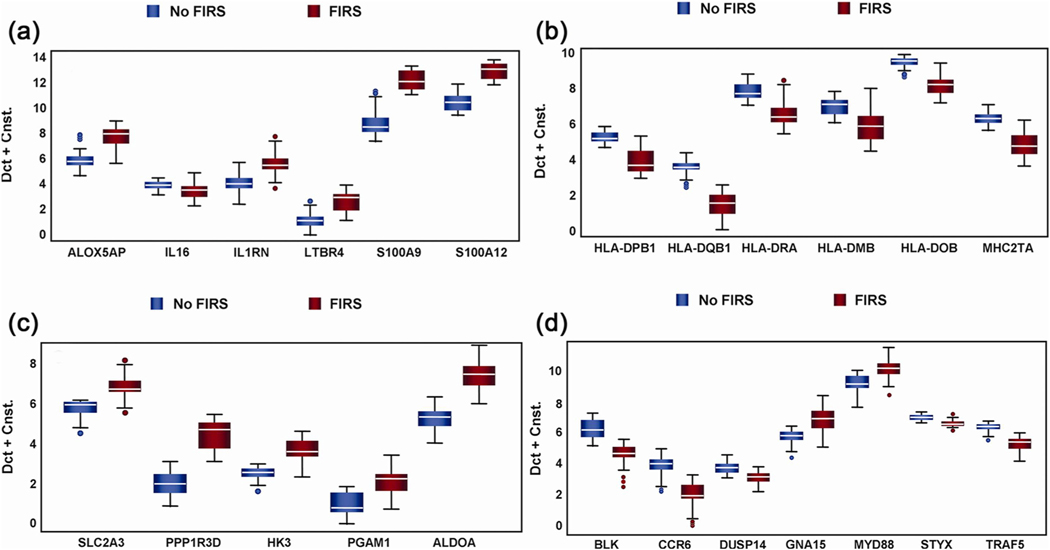
Fig. 9.
Differentially expressed genes in umbilical cord blood according to the presence or absence of fetal inflammatory response syndrome (FIRS). The data were generated by qRT-PCR. Genes were selected after the results of microarray analysis. Altered abundance of genes within ontological categories of immune response and inflammation (a), MHC II receptor activity (b), carbohydrate metabolism (c) and signal transduction (d) was confirmed using qRT-PCR. Box-plots include 50% of the data with the middle line showing the median value; P < 0.05 for all genes is shown when modeled using GEE. Depending on the statistical method used, 93–95% of the genes tested confirmed the change observed by microarray analysis. (Reproduced with permission from Madsen-Bouterse SA, Romero R, Tarca AL et al. The transcriptome of the fetal inflammatory response syndrome. Am J Reprod Immunol 63:73–92, 2010.).