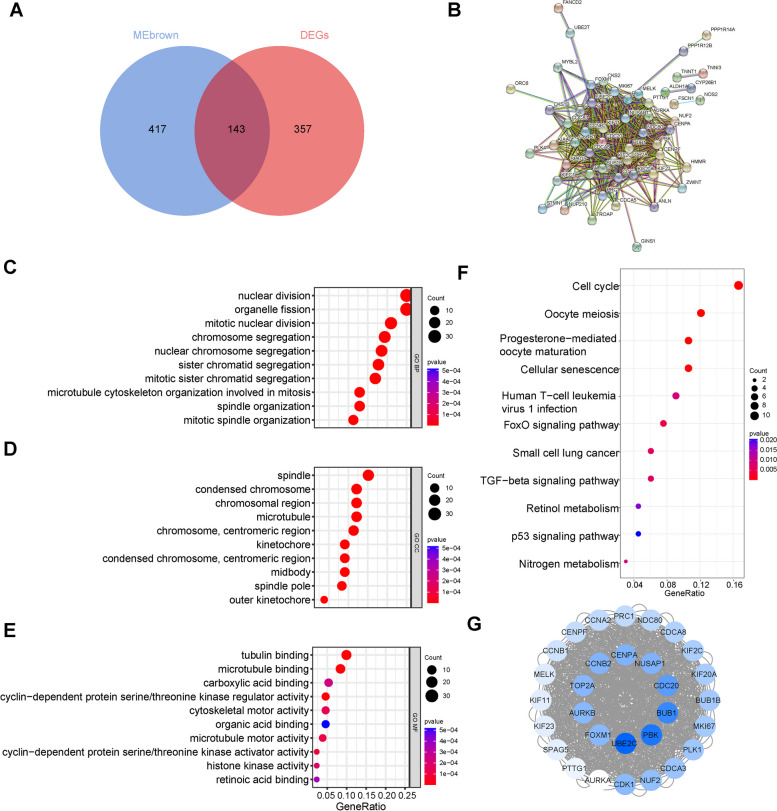
Fig. 4.
Identification of hub genes using a comprehensive strategy. A Venn diagrams of 143 candidate genes obtained through intersecting the genes in brown module and DEGs. B PPI network of 143 candidate genes based on the STRING online database. C The top 10 BP terms of 143 candidate genes. D The top 10 CC terms of 143 candidate genes. E The top 10 MF terms of 143 candidate genes. F The top 11 KEGG pathway enrichment analysis of candidate genes. G Cluster 1 of PPI network genes identified by MCODE from Cytoscape. Blue color represented the downregulated genes and the darker the color was, the greater the |log2 (FC) | was. The top ten genes with the highest |log2 (FC) | were selected as the hub genes. DEGs: differentially expressed genes; PPI: protein–protein network; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; BP: biological process; MF: molecular function; CC: cellular component; FC: fold change; UBE2C: ubiquitin conjugating enzyme E2 C; PBK: PDZ binding kinase; BUB1: BUB1 mitotic checkpoint serine/threonine kinase; CDC20: cell division cycle 20; CENPA: centromere protein A; NUSAP1: nucleolar and spindle associated protein 1; CCNB2: cyclin B2; TOP2A: DNA topoisomerase II alpha; AURKB: aurora kinase B; FOXM1: forkhead box M1