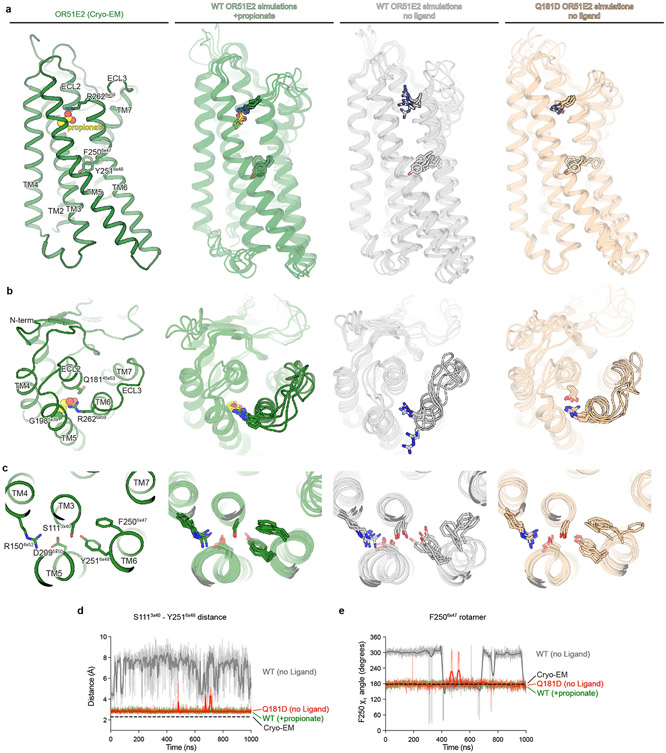
Extended Data Figure 9. Molecular dynamics snapshots of OR51E2.
a) Comparison of cryo-EM structure of propionate-bound OR51E2 with representative snapshots from simulations of WT OR51E2 with propionate, WT OR51E2 without ligand, and Q18145x53D OR51E2 without ligand. Notably, OR51E2 does not transition to the inactive conformation in any of these simulations. b) Close-up views of OR51E2 binding site and ECL3 region in the cryo-EM structure and simulations. In propionate-bound MD simulations of WT OR51E2, R2626x59 persistently forms an ionic interaction with propionate. In simulations of WT OR51E2 with propionate removed, R2626x59 is flexible. Introduction of Asp in position 45x53 (Q18145x53D) stabilizes R2626x59 in an active-like state by a direct ionic interaction. c) Close-up views of OR51E2 connector region shows increased flexibility of WT OR51E2 simulated without propionate. This flexibility is decreased for the Q18145x53D mutant. In a-c, displayed snapshots are the last 1000th ns snapshots from each simulation replicate. d and e) Molecular dynamics trajectories from representative simulations to highlight structural organization of connector region. d) Minimum distance between S1113x40 and Y2516x47 hydroxyl groups is comparable for Q18145x53D and propionate-bound WT OR51E2. e) Rotamer angle of F2506x47is comparable for Q18145x53D and propionate-bound WT OR51E2. Simulations were performed with or without propionate over the course of 1000 ns (see Extended Data Fig. 8 for replicates of simulation trajectories). Thick traces represent smoothed values with an averaging window of 8 nanoseconds; thin traces represent unsmoothed values.