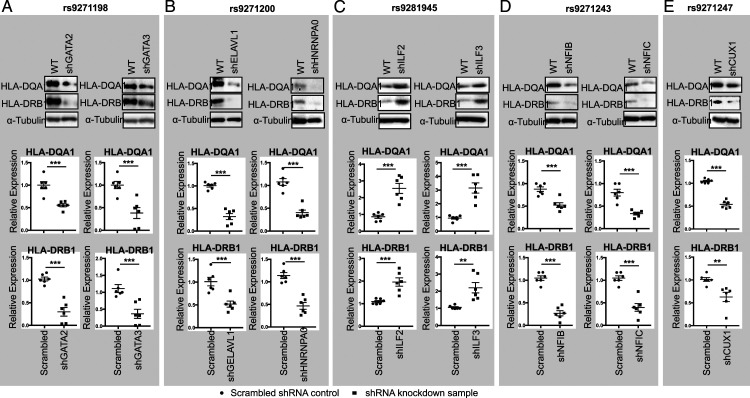
Figure 4.
Western blots (Upper panel) and qPCR (Lower panel) showing the altered expression of HLA-DQA1 and HLA-DRB1 in the GATA2 and GATA3 (A); ELAVL1 and HNRNPA0 (B); ILF2 and ILF3 (C); NFIB and NFIC (D); and CUX1 (E) shRNA knockdown HMC3 cells. Data for Western blots represents three biologically independent experiments (n = 3). Data for qPCR represents the combination of three biologically independent experiments (n = 3), each in duplicate. α-Tubulin: loading control for Western blot and GAPDH was used as control for qPCR. * P < .05; ** P < .001; and *** P, .0001.