Figure 1.
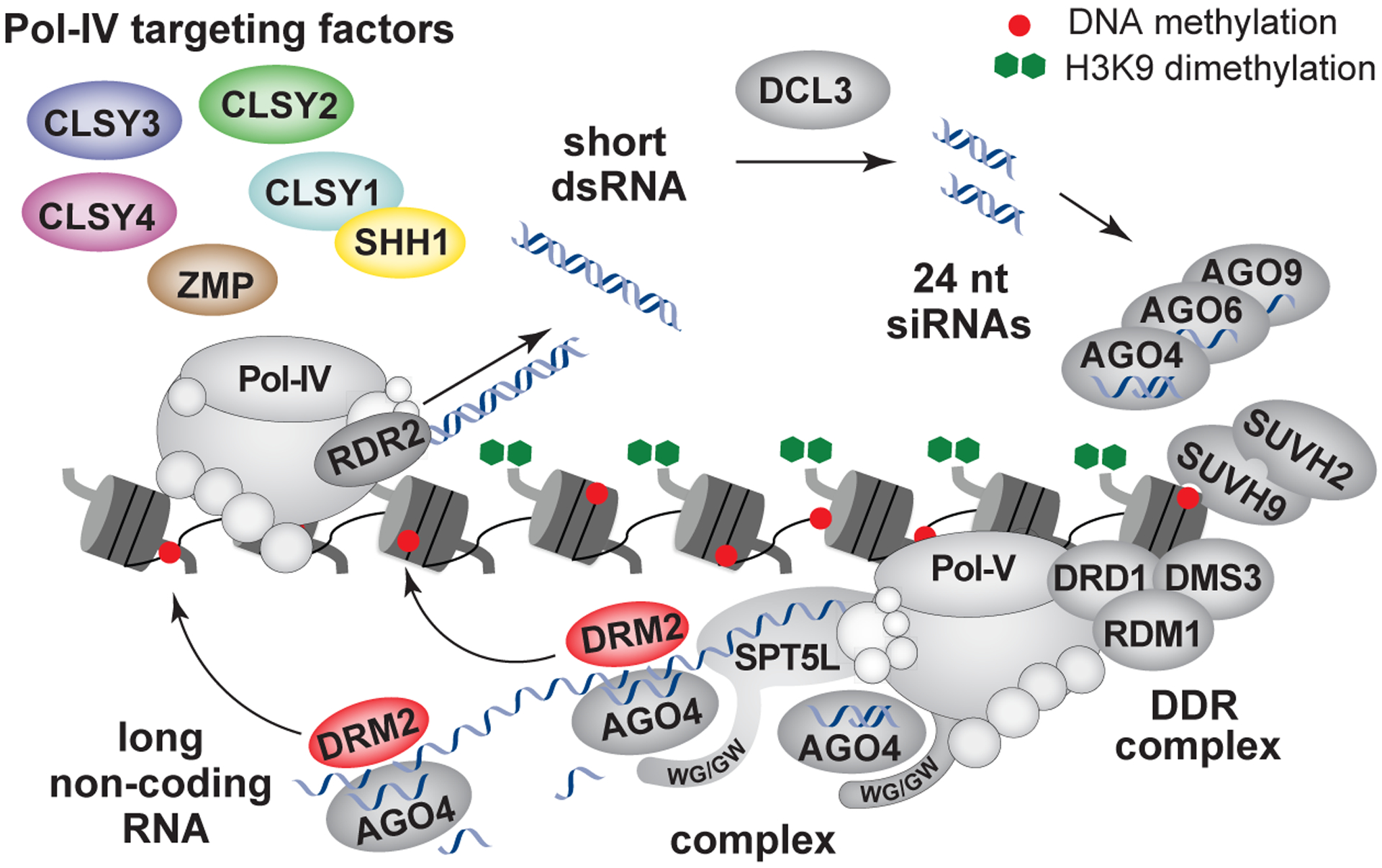
Detailed view of the canonical RdDM pathway, which utilizes two different types of non-coding RNAs to target DNA methylation and silence genes and transposons. Briefly, RdDM is initiated by the Pol-IV polymerase, which interacts with, and is targeted to chromatin by, a number of proteins, including SHH1, all four CLSYs, and ZMP. Pol-IV transcripts are processed co-transcriptionally by RDR2 to generate short double-stranded RNAs (dsRNAs). These RNAs are chopped by DCL3 into 24nt siRNAs, which are loaded into various AGO effector complexes. Pol-V, which is targeted by the DDR complex and two SUVH proteins, generates long non-coding RNAs. AGO effector complexes are recruited to RdDM loci via interactions with the WG/GW motifs present in Pol-V and SPT5L and interactions with the nascent Pol-V transcript. Sliced Pol-V transcripts bound by AGO4 are proposed to promote the association with DRM2 to enable efficient deposition of DNA methylation. Non-canonical RdDM pathways, which generate siRNAs in a Pol-IV independent manner, are reviewed in[43]. The red dots and green octagons represent DNA and histone methylation, respectively.
