Figure 3.
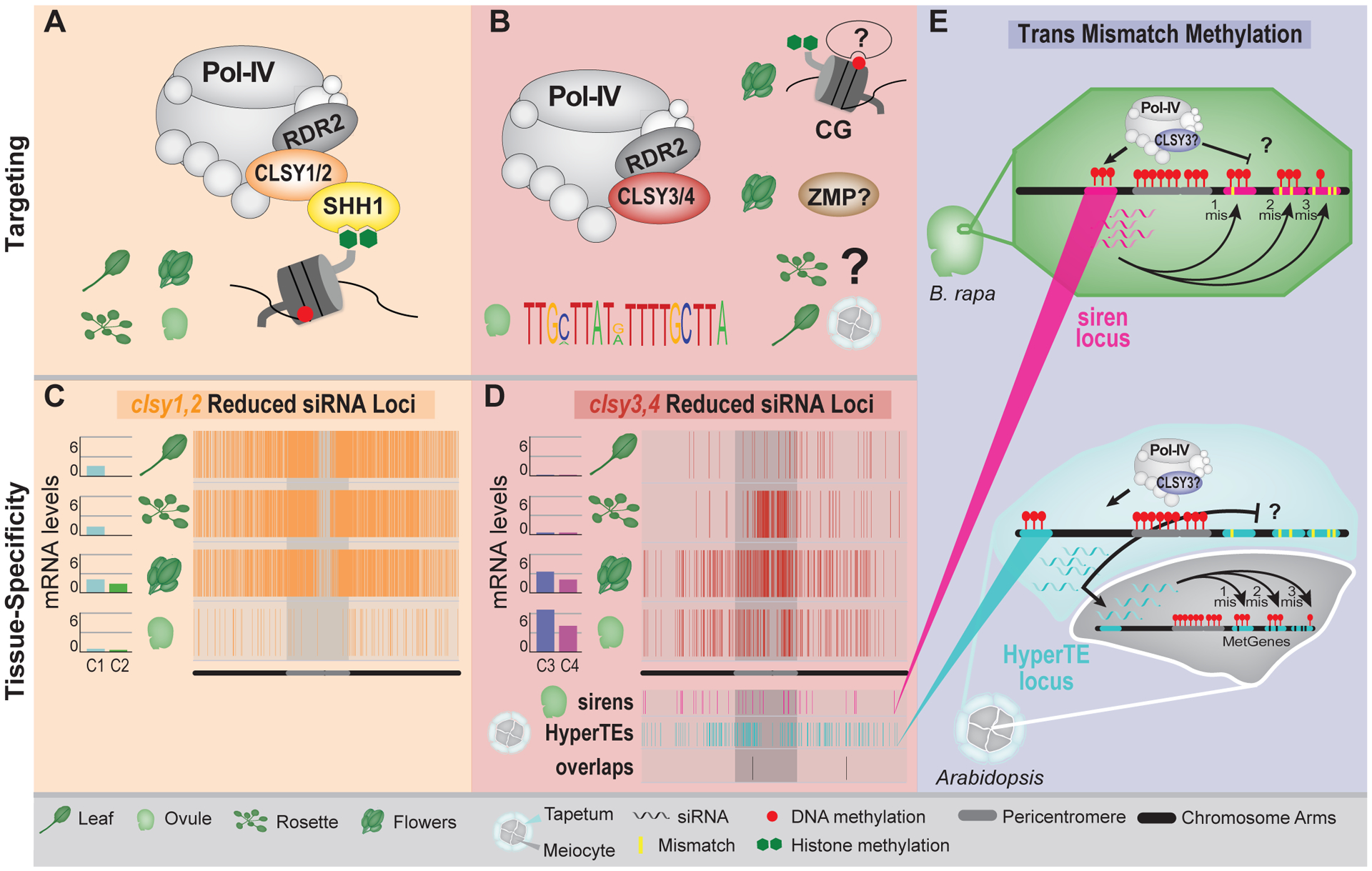
Mechanisms employed by Pol-IV targeting factors to generate epigenetic diversity within and between tissues. (A-B) Cartoons depicting the targeting mechanisms employed by CLSY1 and CLSY2 (A), or CLSY3 and CLSY4 (B), in the indicated tissues. In B, the targeting mechanism(s) in several tissues remain open questions (?). (C and D) CLSY expression levels [log2 (fpkm+1)] and genomic distributions (Chr 1) of reduced siRNA clusters in clsy1,2 (C) or clsy3,4 (D) mutants in the indicated tissues[20]. In D, the distributions of siren loci in ovules and HyperTEs in tapetum cells, as well as the overlaps of these loci, are also included. (E) Cartoons portraying the ability of siRNAs from siren loci to target methylation at genes with imperfect homology in ovule cells (Upper) and the ability of siRNAs from HyperTE loci to move from tapetum cells into neighboring meiocytes to target genes with imperfect homology (Lower). In the former, what blocks Pol-IV from generating perfectly matching siRNAs at the targeted genes remains an open question (?) and in the latter, what blocks the targeting of methylation at imperfectly matching targets in tapetum cells remains an open question (?). The background colors in A, B and E correspond to the Pol-IV targeting factors being discussed.
