Abstract
Animals abandoned their marine niche and successfully adapted to life on land multiple times throughout evolution, providing a rare opportunity to study the mechanisms driving large scale macroevolutionary convergence. However, the genomic factors underlying this process remain largely unknown. Here, we investigate the macroevolutionary dynamics of gene repertoire evolution during repeated transitions out of the sea in mollusks, a lineage that has transitioned to freshwater and terrestrial environments multiple independent times. Through phylogenomics and phylogenetic comparative methods, we examine ∼100 genomic data sets encompassing all major molluskan lineages. We introduce a conceptual framework for identifying and analyzing parallel and convergent evolution at the orthogroup level (groups of genes derived from a single ancestral gene in the species in question) and explore the extent of these mechanisms. Despite deep temporal divergences, we found that parallel expansions of ancient gene families played a major role in facilitating adaptation to nonmarine habitats, highlighting the relevance of the preexisting genomic toolkit in facilitating adaptation to new environments. The expanded functions primarily involve metabolic, osmoregulatory, and defense-related systems. We further found functionally convergent lineage-exclusive gene gains, while family contractions appear to be driven by neutral processes. Also, genomic innovations likely contributed to fuel independent habitat transitions. Overall, our study reveals that various mechanisms of gene repertoire evolution—parallelism, convergence, and innovation—can simultaneously contribute to major evolutionary transitions. Our results provide a genome-wide gene repertoire atlas of molluskan terrestrialization that paves the way toward further understanding the functional and evolutionary bases of this process.
Keywords: comparative genomics, evolutionary rates, terrestrialization, niche shift, mollusc
Significance.
Throughout evolution, animals have made remarkable shifts from marine to land environments, but the genetic changes driving these transitions remain unclear. This study delves into this mystery by focusing on mollusks, which repeatedly moved between marine, freshwater, and land habitats. Analyzing over a hundred molluskan genomes, we discovered ancient gene families gaining new gene copies repeatedly, aiding mollusks’ adaptation to nonmarine habitats, mostly through changes in metabolic, osmoregulatory, and defense systems. Results also suggest a role for genetic novelties and functional convergence, revealing an intricate interplay of genetic factors behind major evolutionary shifts. This comprehensive investigation offers insights into the genomic mechanisms underpinning animals’ successful conquest of new environments, providing a valuable resource for understanding these complex processes.
Introduction
Animal terrestrialization, the evolutionary transition from marine to land-dwelling life-forms, is arguably one of the most dramatic examples of life's adaptability and resilience. Terrestrialization is a complex evolutionary process unfolding along extended periods of time that involves overcoming a large number of physiological and environmental challenges, such as maintaining water balance and osmoregulation, gas exchange, protection from solar radiation, reproduction, and dealing with new pathogens and predators, among others (Shear 1991; Selden 2012). While only a few groups attained fully terrestrial forms (i.e., able to complete every phase of their life cycle outside of water-saturated environments, such as tetrapods or some arthropods), animal lineages managed nonetheless to abandon their marine niche to a greater or lesser extent multiple times across evolution (Little 1983; Selden 2012).
One of the animal phyla where multiple ecological transitions from marine to freshwater and terrestrial environments occur are the mollusks, the second most diverse animal phylum. While most of the extant six to eight mollusk classes remained in the ancestral marine niche (e.g., cephalopods, caudofoveates, solenogasters, chitons, scaphopods, etc.), some groups occupied virtually all aquatic and terrestrial habitats on Earth. Specifically, bivalves and gastropods, the most speciose classes, ventured into different freshwater and marginal (e.g., estuarine) niches, while some gastropods further delved and diversified into terrestrial ones (Romero, Pfenninger, et al. 2016; Vermeij and Watson-Zink 2022), with previous studies suggesting that freshwater and marginal environments likely represented intermediate steps in the gastropod path to land (Strong et al. 2007; Krug et al. 2022; Vermeij and Watson-Zink 2022). In bivalves, the transition from marine to freshwater has occurred multiple times during the evolutionary history of the class, with several orders including both marine and freshwater species (Ponder and Lindberg 2008). Most freshwater species are nevertheless included in the freshwater-only mussel order Unionida (Graf and Cummings 2007). Regarding Gastropoda, multiple independent transitions occurred in three out of the six gastropod subclasses: Caenogastropoda, Heterobranchia, and Neritimorpha (Vermeij and Watson-Zink 2022). Within Heterobranchia, the vast majority of terrestrial gastropods belong to Panpulmonata, a hyperdiverse radiation of gastropods, with the order Stylommatophora comprising most freshwater and the vast majority of terrestrial forms (Krug et al. 2022). The evolutionary replication of the ecological shift out of the sea across mollusks, as described above, provides us with a precious opportunity to study the biological bases of such a broad-scale convergent transition, for which the genomic underpinnings remain mostly unknown.
In order to exit the sea toward freshwater and terrestrial environments, the different mollusk lineages needed to overcome a series of physiological and metabolic challenges associated with coping with environmental stressors. More specifically, adaptations were required in osmoregulation, regulation of temperature, respiration, transpiration, reproduction, development, food acquisition and digestion, metabolism, chemoreception, predatory escape, and pathogen defense (Little 1983). In fact, a series of major morphological novelties and physiological changes have been associated with these adaptations. For instance, the development of an opening in the mantle of Panpulmonata snails (the pneumostome) that controls air passage, reducing the extent to which the pallial cavity is exposed to evaporation and that became contractile in Stylommatophora (Barker 2001; Krug et al. 2022), or the evolution of osmotic and ionic regulation of blood composition in several terrestrial lineages, among others (Barker 2001). Genetic research on mollusk terrestrialization has focused so far on investigating the route taken into land (e.g., Krug et al. 2022) or the exploration of signatures of positive selection in mitochondrial genomes (Romero, Weigand, et al. 2016). Nonetheless, while some of the changes in the gene repertoire (i.e., gene gains, duplications, and losses) driving these adaptations have begun to be explored in this clade (e.g., Sun et al. 2019; Liu et al. 2021), a broad-scale analysis in a large phylogenetic context is still lacking. This is particularly relevant, as gene repertoire evolution has been shown to be an important mode of adaptive evolution (Demuth and Hahn 2009; Chen et al. 2013).
Here we fill in this gap by implementing a genome-wide phylogenomic approach to investigate gene repertoire evolutionary dynamics associated with repeated habitat transitions in the mollusks. For that, we developed a conceptual framework to define and detect parallel and convergent evolution at the level of orthogroup (OG) (i.e., the set of genes derived from a single gene in the last common ancestor of all the species under consideration), and we explored the extent of these mechanisms in ∼100 genomic data sets from virtually all main lineages through a combination of phylogenomics and phylogenetic comparative methods. Our results pave the road toward understanding the genomic underpinnings facilitating mollusk terrestrialization and shed light on the mechanisms driving deep macroevolutionary convergence.
Results and Discussion
Gene Family Expansions and Contractions Shaped the Genomic Landscape of Mollusk Diversification in Nonmarine Environments
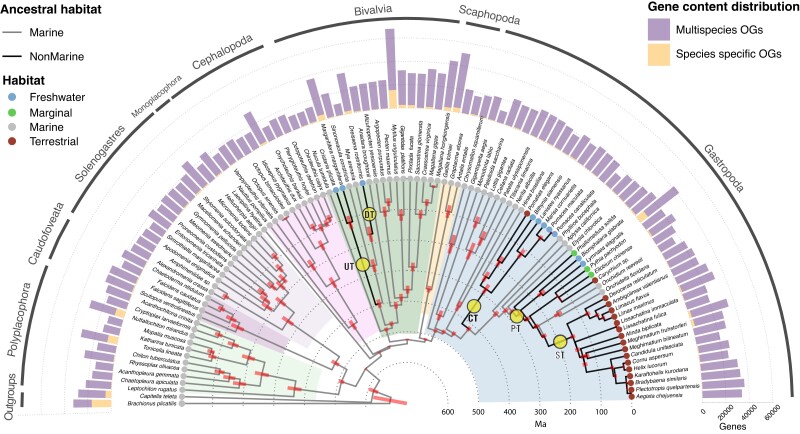
In order to capture the full breath of genomic changes in mollusks facilitating the colonization of new habitats on their march toward land, our analyses focused on the transitions out of marine habitats, including the transition to freshwater environments in bivalves and the transition to both freshwater and terrestrial niches in gastropods. To understand the common genomic underpinnings of these transitions, we first put together a high-quality genomic- and transcriptomic-derived gene repertoire data set (as a proxy of the proteome) comprising 101 mollusks and 2 outgroup species (fig. 1; supplementary table S1, Supplementary Material online). The data set encompasses almost the full diversity of lifestyles, habitats, and morphologies in the clade, thus providing a robust and broad comparative framework required to properly identify gene family changes associated with habitat transitions (Felsenstein 1985). From this data set, we inferred orthology relationships among all genes, resulting in a total of 45,844 OGs (i.e., sets of genes in a sample derived from a common ancestral sequence, OGs hereafter; Altenhoff et al. 2011; Emms and Kelly 2015) containing more than 3 sequences, of which 9,186 were species-specific (fig. 1).
Fig. 1.
Time-calibrated phylogeny for the studied mollusk species. The analyzed data set included 101 mollusks and 2 outgroup species. Bars at nodes indicate 95% confidence intervals of the estimated node ages (see Materials and Methods for further details on molecular dating analyses). Branch intensity indicates the reconstructed habitat state under a maximum-likelihood method (light, marine; dark, nonmarine). Branches connecting nodes with different reconstructed habitats are deemed as “transitional” (circles; bivalve transitions: Unionidae [UT[ and Dreissenidae [DT]. Gastropod transitions: Caenogastropoda [CT] and Panpulmonata [PT]. ST transition involves a fully terrestrial clade and is also included in innovation analyses; see main text). Bars in the external semicircle indicate, for each species, the number of genes in its proteome clustered in species-specific or multiple-species OGs. Only the latter were included in our analyses.
With the aim of pinpointing gene repertoire changes potentially associated with the habitat shift, we implemented a model selection approach based on a stochastic birth–death process of gene family size evolution (Librado and Rozas 2022), to identify OGs that have expanded or contracted at significantly elevated rates in nonmarine lineages compared with marine ones (fig. 1). The application of such a process-based model allowed us to establish a null expectation of background stochastic expansions and contractions across the mollusk tree and against which we can identify gene families in the clades of interest departing from this expectation—that is, that are likely to have evolved under a different evolutionary regime. In this sense and in contrast with hypothesis-driven investigations of well-characterized gene families, our top-down approach can be thought as hypothesis-generating, as even gene families of unknown function could be included in the reduced set of gene families showing a strong evolutionary signal (i.e., a fast evolutionary rate) that may deserve further functional and phylogenetic investigation. This way, our approach for identifying families associated with potential adaptations is more rigorous than methods that solely look for gene content differences between species and in a limited phylogenetic context.
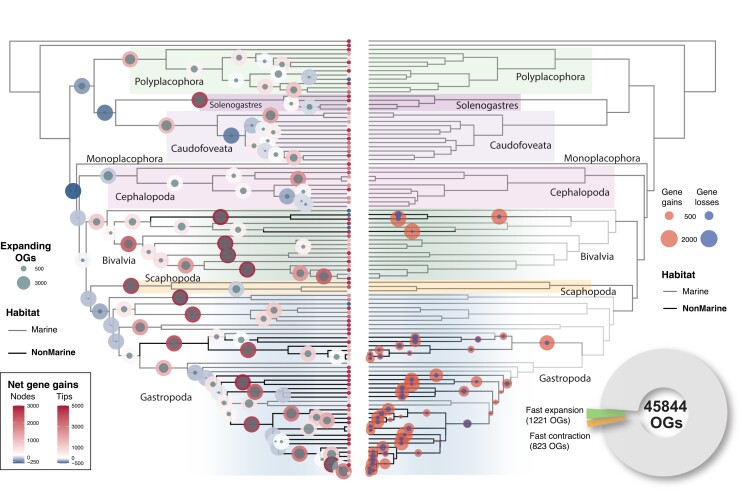
To have a first glimpse at the global repertoire dynamics in the clade, we reconstructed the overall changes across the mollusk tree based on the best-fitting model for each OG (fig. 2, left panel). These results revealed a highly dynamic landscape of expanding and contracting gene families. Notably, repertoire expansions were more prevalent in bivalves and gastropods (the most speciose and ecologically diverse mollusk groups), while cephalopods, caudofoveata, chitons (Polyplacophora), and solenogasters appeared as more genomically quiescent (i.e., smaller repertoire changes). Moreover, while most terminal branches showed repertoire expansions as expected, contractions were concentrated at the origin of main groups. Together, these results suggest that a significant remodeling of the ancestral gene repertoire underlied the emergence of the major mollusk clades, while posterior and heterogeneous lineage-specific expansions provided the genomic material that fueled intraclade diversification.
Fig. 2.
Gene repertoire evolution across the mollusks. Left panel: at each internal branch is indicated the net repertoire changes (i.e., total number of genes gained or lost across all gene families; outer circle) and the number of OGs undergoing expansions (inner circle). At tips, net repertoire changes are shown (different scale for visualization purposes). Right panel: dynamics for those OGs identified as fast-expanding (total gene gains) or fast-contracting (total gene losses) in nonmarine lineages (inset pie chart). Fully labeled versions of these figures are provided in supplementary figures S1 and S2, Supplementary Material online.
Against this background of genomic change, our phylogenetic comparative analyses identified 1,221 and 823 OGs that were either expanding or contracting faster than expected in nonmarine lineages (false discovery rate [FDR]-adjusted P < 0.05; fig. 2, right panel; supplementary tables S2 and S3, Supplementary Material online). These OGs likely evolved under a different evolutionary regime and thus constitute a set of OGs of potential evolutionary relevance for habitat transitions.
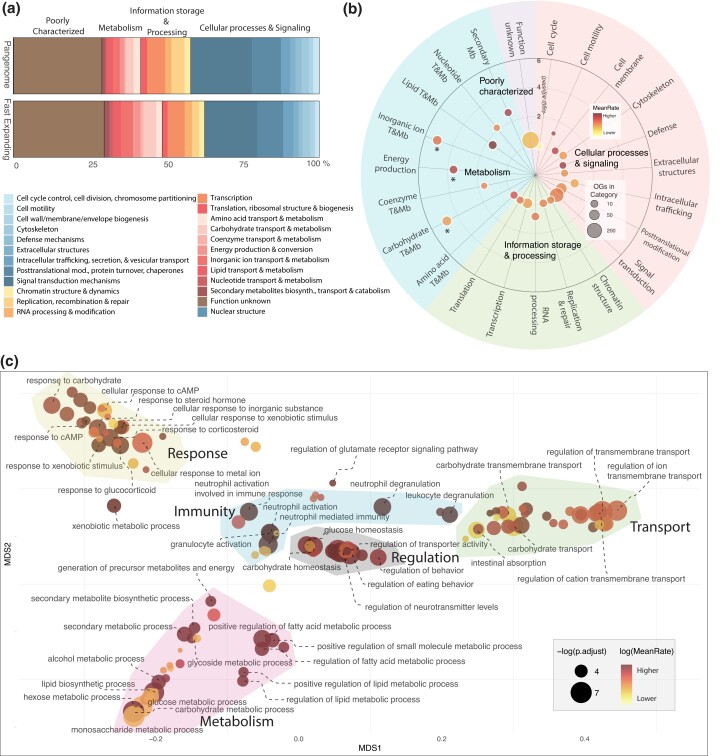
To further explore these OG sets, we investigated their putative functions through evolutionarily based functional annotation and enrichment tests. Our results revealed a contrasting picture between the fast-expanding and fast-contracting OGs. Noticeably, we did not recover any enriched terms for the fast-contracting set (not shown), suggesting that contractions broadly targeted all biological systems more or less randomly. Conversely, an examination of the high-level functional categories from the Cluster of Orthologous Genes (COG) database (Tatusov et al. 2000) revealed that metabolism, as a broad category, is overrepresented in the set of fast-expanding OGs (fig. 3a). More specifically, the narrower COG metabolism categories of energy production and conversion and transport and metabolism of carbohydrates and inorganic ions were significantly enriched (fig. 3b; supplementary table S4, Supplementary Material online; P < 0.05). The latter category could suggest a significant role for gene family expansions in meeting osmoregulatory and water balance needs in nonmarine environments, while fast expansions of gene families in the first two categories is consistent with potential dietary shifts and with the expectation that when facing multiple environmental stressors, organisms need to adapt by allocating significant energy away from functions like growth or reproduction to maintain internal homeostasis (Sokolova et al. 2012). Further supporting this, lipid transport and metabolism appear as the category with the fastest mean rate of OG expansion, despite not being significantly enriched (fig. 3b; supplementary table S4, Supplementary Material online). Similarly, defense mechanisms and secondary metabolite biosynthesis, transport, and catabolism were also the COG categories associated with the highest rates of OG expansion (fig. 3b), likely associated with the challenges posed by environmental stressors (e.g., temperature and oxygen fluctuations), new pathogens (e.g., parasites), and xenobiotics (e.g., dietary toxins) encountered out of the sea.
Fig. 3.
Functional characterization of fast-evolving gene families in nonmarine mollusk clades. a) COG categories proportions in the fast-expanding OG set, compared with the mollusk pangenome annotation. b) Enrichment of COG terms in the fast-expanding OG set, with the mean expansion rates of the OGs annotated to the term. Asterisks indicate significantly enriched terms (adjusted P < 0.05) c) GO terms semantic space showing enriched terms in the fast-expanding OG set, with their associated mean expansion rates (as in b). Colored polygons describing broad categories were drawn by hand.
Overall, the functional interrogation of the fast-evolving OGs suggests a contrasting picture. On the one hand, gene gains provided the genomic material that likely fueled the transition and diversification into nonmarine forms, targeting several biological systems that allowed them to tolerate and evade physiologically stressful conditions and to adapt and thrive in massively different ecological and environmental niches compared with the ancestral condition. On the other hand, gene losses in fast-contracting OGs were not associated with specific functions, likely representing the action of more neutral processes (i.e., relaxation of selection and drift) across all biological systems (Albalat and Cañestro 2016; Ohno 1985). This picture is further refined by enrichment tests for gene ontology (GO) terms, which recovered 117 significantly overrepresented terms in fast-expanding OGs (P < 0.05; fig. 3c; supplementary table S4, Supplementary Material online), among which carbohydrate-related processes (e.g., metabolism), transmembrane transport, immunity, and xenobiotics-related terms stand out (fig. 3c). As a discussion of each fast-expanding OG would be impossible, we here focus on discussing with more detail some of those that we deem as more relevant in the light of previous work based on functionally characterized genes in mollusk species and other invertebrates, namely, carbohydrate metabolism, transmembrane transport, immune system, and stress response.
Carbohydrate Metabolism
The fast-expanding OGs annotated to carbohydrate metabolism-related terms included 10 glycosyl hydrolases (GH; supplementary table S5, Supplementary Material online), key enzymes involved in the degradation of complex carbohydrates (Naumoff 2011). An increased repertoire of these enzymes could have facilitated the dietary shift associated with the habitat transitions, as gastropods and bivalves likely started relying more heavily on freshwater algae and plant matter for energy, which differ from marine algae in their polysaccharide composition (Popper et al. 2011). Consistent with this interpretation, a previous study on seven marine mollusks identified at least 26 GH families in the clade, of which two (GH9 and GH10) were reported as expanded in algae-feeding species compared with carnivorous species (Wang et al. 2020). Here, we similarly recovered two fast-expanding OGs belonging to GH9, cellulose-degrading enzymes of ancient origin found across metazoans (Davison and Blaxter 2005). Cellulose is particularly abundant in green algae and plants (Popper et al. 2011), which suggests that these enzymes might be of particular adaptive relevance for habitat transitions. In fact, another cellulase, GH5, appears also among the fast-expanding OGs (supplementary table S5, Supplementary Material online). Interestingly, Sun et al. (2019) have previously found an expansion of another cellulase family (GH10) in ampullariids (a group of freshwater snails including several highly invasive Pomacea species in the Caenogastropoda superorder), which was also highly expressed in the digestive gland, suggesting that this may have facilitated the evolution of the trophic versatility seen in the clade.
Another example of a fast-expanding OG of potential relevance related to carbohydrate metabolism and energetic homeostasis, is glycogenin (GYG1), the initiating enzyme of glycogen synthesis (i.e., the main form of glucose storage in animals; Roach et al. 2012). Interestingly, in a phylogenetic reconstruction of this OG (supplementary fig. S3, Supplementary Material online), we observed a highly divergent clade formed by terrestrial and freshwater snails from the Panpulmonata and Caenogastropoda clades. Intriguingly, these snails have been previously reported as being unique among animals in that they produce, in addition to glycogen, large quantities of a polysaccharide of galactose (galactogen) found exclusively in the female albumen gland and the eggs (Livingstone and de Zwaan 1983), with only the embryos and hatchlings being able to catabolize it (Giglio et al. 2016). This might suggest an antipredatory role for galactogen, as it would reduce the nutritional value of the conspicuous egg clutches that some of these species lay outside of the water (Giglio et al. 2016). Galactogen biosynthesis pathway, while not completely understood, is likely closely related to that of glycogen, and thus the expansion of the GYG1 OG could be related not only to satisfying energy storage needs in the new habitats (e.g., aestivation in gastropods) but also with the acquisition of novel functions by the extra gene copies generated during duplication, fueling new key adaptations (i.e., a genomic exaptation; Gould and Vrba 1982).
Transmembrane Transport
Another category potentially of key relevance for habitat transitions is transmembrane transport, as it is tightly linked to nutrient absorption, detoxification, osmoregulation, and shell formation. We found representatives of several transporter families among the fast-expanding OGs, for example, ATP-binding cassette (ABC) transporters, aquaporins (AQP), and solute carriers (SLC), among others (fig. 3c; supplementary table S6, Supplementary Material online). ABC transporters function mostly as efflux proteins involved in lipid metabolism and xenobiotic detoxification, with the latter being particularly relevant in filter feeders as bivalves (Luckenbach and Epel 2008), and more generally in herbivores, in which they contribute to dealing with plant chemical defenses (Sorensen and Dearing 2006). Noticeably, ABC transporters were reported as upregulated under stress conditions in the freshwater snail Pomacea canaliculata (Liu et al. 2018), while Sun et al. (2017) reported an expanded repertoire in the deep-sea mussel Bathymodiolus platifrons, which thrives in the highly toxic environments around hydrothermal vents. Our results further highlight the potential relevance of these transporters, likely in the light of new detoxification needs associated with dietary shifts during habitat transitions.
We also recovered two aquaporin families (AQP3 and AQP8) as fast-expanding in nonmarine mollusks, which are pore-forming membrane proteins that facilitate the transport of water and other small solutes and are heavily involved in osmoregulation (Kruse et al. 2006). This result is in line with a previous study showing repeated expansions of aquaporin genes in association with transitions out of the sea in mollusks and other animal phyla (Martínez-Redondo et al. 2023). Finally, our analysis recovered at least 19 SLC fast-expanding OGs belonging to 18 different families (supplementary table S6, Supplementary Material online). SLCs constitute a broad class of transporter proteins grouped in more than 60 families that regulate transport of a multitude of different molecules over cell membranes, having equally diverse physiological roles (Pizzagalli et al. 2021). Noticeably, among the fast-expanding SLC, we identified members of the SLC4, SLC12, and SLC26 families, which were previously found to be upregulated in the shells of several bivalve species and hypothesized as being part of a bivalve genetic “biomineralization toolbox” involved in shell formation (Yarra et al. 2021). SLC4 is a bicarbonate transporter also involved in dentition formation in mice (Lacruz et al. 2010) and sea urchin (Hu et al. 2018) and coral calcification (Bhattacharya et al. 2016), and its fast expansion in nonmarine mollusks could be related to the need to adjust the availability of bicarbonate ions (e.g., for pH regulation) for proper calcium deposition in the context of the different physicochemical properties of freshwater and terrestrial environments compared with marine ones. Similarly, SLC12, a family of cation-coupled chloride transporters, has a strong osmoregulatory role in humans (Arroyo et al. 2013), and it is involved in sea urchin calcification (Basse et al. 2015). Another noteworthy SLC identified as expanded in our study is SLC34A2. This is a sodium phosphate cotransporter that has purportedly played a role in vertebrate terrestrialization, as it is highly expressed in the lungs of lungfish and tetrapods where it has a key role in the recycling of pulmonary surfactant (Wang et al. 2021), a complex mix of phospholipids and proteins whose function is to prevent the collapse of gas-holding structures. Strikingly, a surfactant with a very similar composition to that found in air-breathing vertebrates has been described in the lung cavity of the pulmonate terrestrial snail Helix aspersa (Daniels et al. 1999), suggesting that the surfactant system might have very deep evolutionary roots (Daniels and Orgeig 2001). Although further work is needed to determine the specific role of SLC34A2 in mollusks, our results open up the intriguing possibility of a shared genomic basis for the evolution of air breathing across extremely large evolutionary scales.
Immune System and Stress Response
We recovered five C-type lectin OGs (supplementary table S7, Supplementary Material online), which are core innate immunity proteins that mediate microbial pathogen recognition through binding different carbohydrate moieties in their surfaces (Cambi et al. 2005). In this sense, a fast expansion of lectin genes may have provided an increased ability to recognize newly encountered pathogens outside marine environments. Of note, the genome of Nautilus pompilius exhibits a large number of C-type lectins compared with other cephalopods (Zhang et al. 2021), which suggest that expansions of these genes could provide a readily available evolutionary solution to immune challenges in mollusks. Also, C-type lectins were found to be upregulated under abiotic stress in the Pacific oyster (Crassostrea gigas), highlighting potential interactions between stress and immune responses (Zhang et al. 2015). Another set of fast-expanding OGs associated with immunity and defense are those associated with the mucus system, which involves the secretion of highly glycosylated proteins (e.g., mucins; McShane et al. 2021). In this regard, we detected several OGs involved in protein glycosylation (e.g., glycosaminoglycan, and O- and N-glycan biosynthesis; supplementary table S8, Supplementary Material online), as well as several mucin OGs, which suggests that the mucus system may have played a significant role during mollusks’ habitat transitions. Similarly, Liu et al. (2021) described an expansion of mucus-related gene families in panpulmonate gastropods (but also in Cephalopods), with a noticeable expansion of mucin genes in the terrestrial Stylommatophora clade.
Other stress and defense-related fast-expanding OGs are those belonging to the heat shock protein (HSP), the glutathione S-transferases (GSTs), and the cytochrome P450 (CYP) families (supplementary table S9, Supplementary Material online). HSPs are highly conserved molecular chaperones key for cellular homeostasis, with some members having a markedly increased expression under different stress conditions (e.g., HSP70; Sørensen et al. 2003). In this sense, Zhang et al. (2012) demonstrated a genomic expansion of HSP70 proteins in C. gigas and a many-fold increase in their expression under heat and other stressors, with a similar response observed in the freshwater snail P. canaliculata (Liu et al. 2018). Here, we similarly recovered two HSP70 fast-expanding OGs in nonmarine mollusks, highlighting the evolutionary universality of this system. Similarly, GSTs are a fundamental part of the detoxification and antioxidant systems, dealing with numerous by-products of oxidative stress and the CYP detoxification pathway (Strange et al. 2001). Here we detected four fast-expanding GSTs families, suggesting that these enzymes likely played an important role in conferring resistance to stressful environmental conditions (e.g., hypo- and hyperoxia) during habitat transitions. Finally, we found four fast-expanding CYP families, key enzymes in the metabolism of xenobiotics, and the biosynthesis of hormones that are relevant for the stress response (e.g., glucocorticoids; Denisov et al. 2005), a result in line with Liu et al. (2018) who found an expanded repertoire of CYPs and upregulated expression under stress in P. canaliculata.
Parallel Gene Repertoire Expansions During Transitions out of the Marine Niche in Bivalves and Gastropod Mollusks
Our rate-modeling approach allowed us to identify OGs that were overall fast-evolving along the evolutionary diversification of nonmarine clades, encompassing all stages and degrees of adaptation to different nonmarine habitats (fig. 1). To further refine our understanding of the abandoning of the marine niche, we next focused on the repertoire changes in the fast-evolving OGs occurring specifically on the internal branches of the tree where we can confidently reconstruct independent transitions out of the sea: two in the bivalves (Unionida and Dreissenidae) and two in gastropods (Caenogastropoda and Panpulmonata; fig. 1). These replicated transitions provide an excellent opportunity to robustly test for associations of gene gains with habitat shifts, as a signal of repeated parallel evolution would constitute strong evidence for the role of these changes in adaptation (Losos 2011; Stayton 2015; Gompel and Prud’homme 2009).
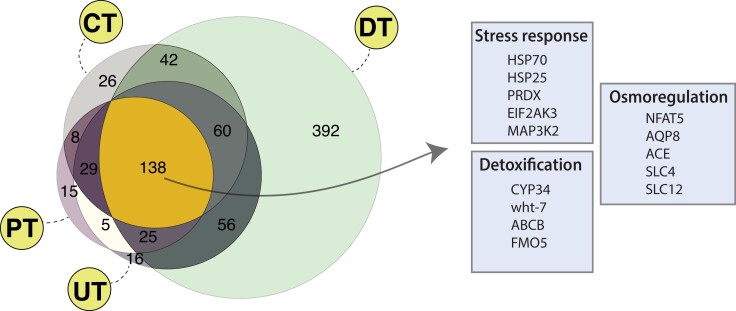
Our reconstruction of OG size changes in the fast-expanding set revealed that out of 869 OGs that had net gene gains in the four transition branches, 138 were shared among them (fig. 4; supplementary table S10, Supplementary Material online), which we interpret and define as evidence of parallel evolution (i.e., the same OGs underwent similar expansions in the four transition branches). Functional exploration of these parallel OG expansions further highlighted the potential adaptive relevance of the new genetic material: the two COG categories that are significantly enriched (supplementary table S11, Supplementary Material online) are “secondary metabolite biosynthesis, transport and catabolism” and “inorganic ions transport and metabolism,” which, as discussed previously, are associated to potential key functions for habitat transitions, such as detoxification and osmoregulation. In this sense, among the enriched GO terms (supplementary table S11, Supplementary Material online), those related to xenobiotics (e.g., cellular response to xenobiotic stimulus), ion transport (e.g., regulation of ion transmembrane transporter activity), and water balance (e.g., cellular hyperosmotic response) appear repeatedly. More specifically, related to detoxification, we found two ABC transporter families and one CYP (see above for discussion), as well as a flavin-containing monooxygenase (FMO; involved in phase 1 biotransformation of xenocompounds, and likely found across metazoans; Goldstone et al. 2006) that expanded in parallel (supplementary table S12, Supplementary Material online).
Fig. 4.
Parallel and exclusive OG expansions in the four out-of-the-sea transition lineages. Numbers in the Euler diagram (left) indicate the number of shared and exclusively expanded OGs among the different lineages. Parallel expansions in the four transition branches (arrow) involve families and functions likely relevant for the habitat transitions. Some examples are shown on the right (see text for discussion). Bivalve transitions: Unionidae (UT) and Dreissenidae (DT). Gastropod transitions: Caenogastropoda (CT) and Panpulmonata (PT).
Related to water and ion homeostasis, we found AQP8 and five SLC OGs that expanded in parallel, including the SLC4 family likely involved in shell formation (see above). Interestingly, we also found parallel expansions of the NFAT5 transcription factors, which have a demonstrated role in the osmotic stress response in mammals (Cheung and Ko 2013), insects (Keyser et al. 2007), and teleosts (Lorgen et al. 2017), where it notably induces the expression of several SLCs, AQP, and HSP70 genes. Although it has been little studied in mollusks, NFAT5 genes were identified in the pearl oyster (Pinctada fucata) genome, where they were shown to be activated upon immune challenge (Huang et al. 2015). Also potentially related to osmoregulation, we found parallel expansions in the angiotensin-converting enzyme (ACE) family, a key component of the renin–angiotensin hormonal system that, among other functions (e.g., immune), controls fluid volume in the body (Salzet et al. 2001).
Lastly, in addition to the previously discussed HSP70, we identified other parallel expansions in the stress response-related gene families EIF2 kinases, and peroxiredoxins (PRDX). EIF2 kinases include at least four genes (EIF2AK1-4) involved in the downregulation of protein synthesis in response to various forms of cellular stress (Hinnebusch 1994), with EIF2AK3 (or PERK) being one of the three molecular sensors that control the activation of the unfolded protein response (Korennykh and Walter 2012). PRDX or Prx form a large family of antioxidant enzymes plays a fundamental role in protecting cells from oxidative damage (Wood et al. 2003) and, suggestively, has been found to be upregulated in response to stress in the snail P. canaliculata as well as in the unionid bivalve Anodonta woodiana, both inhabiting freshwater environments (Liu et al. 2018; Xia et al. 2018).
Overall, our analyses of the parallel gene repertoire changes on transition branches highlight the repeated use, despite deep temporal and biological divergences, of preexisting genomic material (i.e., ancient gene families) involved in conserved defense-related pathways as a mechanism to fuel adaptations in the face of a similar both ecological challenge and opportunity.
Functional Convergence Underlies Nonparallel Gene Repertoire Expansions During Transitions Out of the Sea
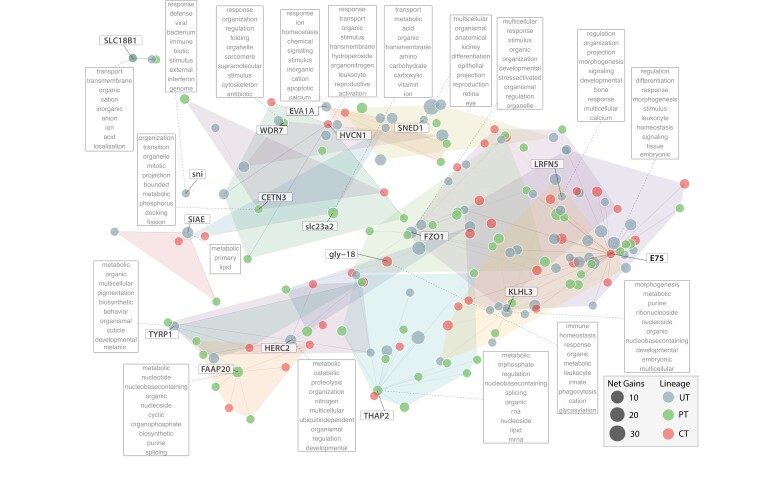
While the parallel expansion of gene families during the replicated transitions out of the sea is strongly suggestive of their adaptive relevance, the role played by lineage-exclusive expansions is harder to pinpoint. However, as high-level biological functions can be fulfilled through alternative genetic mechanisms or pathways (e.g., acquisition of energy through lipid or carbohydrate metabolism), it may be expected for evolution to lead, when repeatedly confronted with an ecological challenge, to convergent functional solutions despite differences in their genomic underpinnings (Rosenblum et al. 2014). We next tested this hypothesis by investigating the potential functional similarities among the OGs that were expanded exclusively in each of three ancestral mollusk transitional branches (the Dreissenidae transition was excluded as it is represented in our sample by a single species, and thus, it is not possible to disentangle from all lineage-exclusive expansions those that are ancestral and potentially linked to the habitat transition from those that are recent and species-specific). To do so, we developed a methodological framework to group the exclusively expanded OGs in functional clusters based on their GO annotations, which we represent in what we name as constellation plots (see Materials and Methods). Our results recovered 18 functional clusters, most of which are composed of OGs exclusively expanded in two or three transition branches (fig. 5; supplementary table S13, Supplementary Material online). For example, the cluster that has HVCN1 as the exemplar OG (i.e., the prototypical or most representative observation of each cluster; see Materials and Methods), is composed of OGs broadly related to immunity and defense (supplementary table S13, Supplementary Material online). HVCN1 is a voltage-gated proton channel that has been shown to be involved in production of reactive oxygen species linked to defense against parasitic infections in the freshwater snail Biomphalaria glabrata (Wright et al. 2017). Another member of this cluster, MGST1, is a microsomal GST (see above), enzymes with broad antioxidant and defense functions, while QPCT (a glutaminyl-peptide cyclotransferase) has been involved in posttranslational modification of snail toxins (Buczek et al. 2005; Gorson et al. 2015). As another example, the cluster defined by gly-18 involves functions likely related to the mucus system. Specifically, it is composed of OGs involved in glycan biosynthesis (gly-18 and CHST1), a potentially mucin-related gene (MFGE8), and TMEM165 which in humans is associated to a congenital disorder of glycosylation (Foulquier et al. 2012). Other convergent clusters of OGs appear to target functions such as pigmentation (cluster defined by TYRP1, a tyrosinase that has been involved in shell formation and pigmentation in oysters; Zhu et al. 2021), protein ubiquitination and posttranslational modification (cluster HERC2), or the regulation of gene expression (cluster THAP2), among others. Overall, these results are suggestive of a significant level of functional convergence among these lineages, highlighting that changes in similar high-level biological processes during the repeated transition out of the sea were in part achieved through different genomic routes. In this sense, this may also suggest that genetic constraints did not play a determinant role in limiting (e.g., through pleiotropy or a lack of genetic diversity) or facilitating (e.g., through reusing tightly linked coadapted gene complexes) molluskan adaptation to nonmarine habitats (Blount et al. 2018; Connallon and Hall 2018).
Fig. 5.
Functional convergence among exclusively expanded OGs in transition lineages. The constellation plot shows the GO semantic space for the 165 exclusively expanded OGs in the transitional ancestral branches. Each point represents an OG, filled according to the lineage where it expanded, with size proportional to the number of net gene gains in the branch. Polygons (colors are arbitrary and for visualization purposes only) delimit each functional cluster as obtained by applying the Affinity Propagation clustering algorithm to the pairwise similarity matrix among OGs (see Materials and Methods). Bold labels indicate, for each cluster, the “exemplar” OG putative human (or other model organisms) ortholog gene name. The word lists inside boxes (dotted lines) are the 10 most enriched words in the combined GO annotations of the OGs in the cluster and provide a broad summary of its associated biological processes.
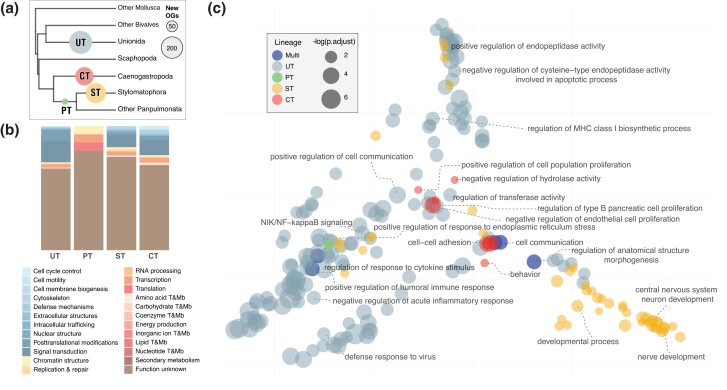
Genomic Innovations Further Potentially Facilitated the Transition out of the sea and into Terrestrial Environments
Our previous analyses revealed how parallel and convergent expansions of preexisting gene families likely facilitated mollusk transitions out of the marine niche. However, it is also known that genomic innovations can be important sources of new functional and phenotypic traits (Kaessmann 2010; Chen et al. 2013). Thus, we lastly identified innovations (here, OGs reconstructed as originating on a given internal branch of the tree, and not present in other branches of the tree other than its descendants, independently of their origination mechanism) during main habitat transitions (Dreissenidae was excluded as above) and investigated their potential relevance in enabling adaptation through functional characterization. Our reconstructions recovered a significant number of innovations occurring during the parallel transitions out of the sea (fig. 6; supplementary table S10, Supplementary Material online). Noticeably, only 15 OGs arose during the Panpulmonata transition compared with the 142 in the other gastropod transition (Caenogastropoda). Given this comparatively low number of innovations during the Panpulmonata transition out of the sea, we also examined innovations occurring later in time in this clade during the transition into terrestriality of the Stylommatophora (fig. 1), a highly diverse clade of snails and slugs comprising most terrestrial species (represented in our tree by species from the Sigmurethra–Orthurethra groups). We found 173 new OGs arising in this branch (fig. 6; supplementary table S10, Supplementary Material online), which coincides with evidence of a whole-genome duplication event taking place at the origins of this group (Hallinan and Lindberg 2011; Liu et al. 2021). This suggests that significant genomic reshaping characterized not only by whole genomic duplication but also through the arisal of new genes may underlie the evolutionary origin and ecological specialization of this clade of mollusks.
Fig. 6.
Genomic innovations in transition lineages. a) Schematic mollusk tree indicating the number of new OGs in each transitional branch (see fig. 1 for a full phylogenetic context). b) COG categories and their proportions annotated to the novel OGs in each transition. (T&MB: transport and metabolism). c) Distribution in the semantic space of enriched GO terms for each transition branch. UT, Unionidae bivalve transition; gastropods: CT: Caenogastropoda; PT: Panpulmonata; ST: Stylommatophora; Multi: Enriched GO terms in more than one lineage.
The functional exploration of the detected innovations was hindered by the low proportion of annotated OGs (<30%), which suggest that most genes emerging in these lineages have novel functions and domains. Interestingly, previous studies revealed that across mollusks, many of the proteins secreted by the mantle during shell formation are the product of lineage-specific genetic novelties, having repetitive, low complexity domains as dominant features (McDougall et al. 2013; Aguilera et al. 2017). Among the new OGs that did have GO annotations, enrichment tests indicated a prevalence of different functions in each lineage (fig. 6). For instance, in the bivalve transition (Unionida), defense- and immune response-related functions were the most enriched, while in the Caenogastropoda transition, terms were mostly related to cell–cell adhesion, pointing to the potential role of the new genes in developmental processes or signal transduction to detect and respond to changes in their surroundings. In the Panpulmonata transition, the single enriched term recovered was related to endoplasmic reticulum stress and unfolded protein response. Finally, innovations in the basal Stylommatophora branch are enriched in functions related to the development of brain and nervous system structures. Overall, while further studies are needed to better characterize these genomic novelties, our results suggest that they could have played a significant role during habitat transitions in the mollusks, highlighting evolution's creative potential as a driving force in evolutionary transitions.
Conclusions
Evolutionary transitions underlying drastic ecological shifts constitute magnificent demonstrations of life's capacity to evolve. However, their genomic underpinnings are mostly unknown, particularly at a macroevolutionary scale. Here, we investigated the dynamics of gene repertoire evolution during replicated transitions out of the sea in mollusks. Using a large genomic data set and phylogenetic comparative methods, we show that expansions of ancient gene families played a major role in facilitating adaptation to nonmarine habitats in gastropods and bivalves. These expansions targeted several biological systems, such as those related to meeting energetic demands (e.g., carbohydrate metabolism) and particularly those related to protection from environmental stressors, such as detoxification, immune, and osmoregulation pathways. Our results highlight the evolutionary potential of the preexisting genomic toolkit, revealed by parallel gene family expansions across the different lineages despite their deep biological and temporal divergences. Moreover, we find a significant functional convergence targeting relevant functions for the habitat shift, underscoring evolution's capacity to “find its way” despite potential constraints, by reshaping common biological systems through alternative genomic pathways (Jacob 1977). On the other hand, we find evidence that gene family contractions did not target specific function, and thus may be mostly driven by processes such as drift, reflecting a global reshaping of most biological systems. Their potential role in driving mollusk terrestrialization should be the focus of future work. Our results also indicate that genomic innovations (i.e., new gene families) also likely contributed to evolutionary independent habitat transitions.
Biologists have discussed and differently emphasized the role of innovations and convergent and parallel changes in driving diversification and adaptation at macroevolutionary scales (Losos 2011; Blount et al. 2018; Erwin 2015). Collectively, our results highlight that, in fact, all these mechanisms can be at play at the genomic level during major evolutionary transitions. Finally, we provide a genome-wide gene repertoire atlas of the molluskan terrestrialization process that will be of great value for future studies looking to further understand its functional and mechanistic bases and the role of specific biological processes in driving adaptation.
Materials and Methods
Taxon Sampling and Data
A total of 101 molluskan genomes and transcriptomes representing all main orders were retrieved from public databases, while specifically targeting mollusk species related to marine-freshwater or marine-terrestrial transitions (available in public databases as of March 2021). Although such data were available for more species potentially relevant to understanding habitat transitions than the ones included in this study, some of them did not meet the minimum quality criterion (i.e., BUSCO score > 80%) and therefore were not included in this study to avoid any artifacts. We also included several species from mollusk clades that are exclusively marine, representing virtually all mollusk classes. Including a wide range of mollusk species regardless of their habitat is of importance to provide a strong phylogenetic context to our analyses, which increases the robustness of the association between habitat transitions and gene repertoire changes. Finally, in this initial data set, 16 outgroup species were included to provide a broad phylogenetic context for gene family delimitation (supplementary table S1, Supplementary Material online).
Habitat data were retrieved from the primary literature and online records (e.g., WoRMS database; WoRMS Editorial Board 2022). Some marine species inhabiting intertidal or estuarine zones might present adaptations to temporally tolerate desiccation, salinity changes, or exposure to direct sunlight (e.g., oysters or chitons). Others, such as Onchidella, inhabit intertidal marine habitats despite being air-breathing pulmonate gastropods. However, we took a conservative approach and only included in the nonmarine group species that inhabit most of the time in conditions that are fundamentally different from the ancestral marine niche. Although by doing so, some adaptations could be potentially overlooked, it avoids a fine-grained habitat classification that in many cases would be difficult given the essentially continuous nature of ecological niches in the mollusks. Moreover, by focusing on clade-wise adaptations and two habitat categories, we mostly avoid the problem of assigning ancestral states of a multicategory trait on the internal branches of the tree, a requisite for phylogeny-based association methods. Such reconstructions are riddled with uncertainty and are influenced by species sampling; thus, relying on them could erase or create artifactual evolutionary signals that would compromise our analyses.
Data Preprocessing
Genomic data processing fundamentally followed the pipeline described in (Fernández et al. 2022). Basically, after trimming of adapters with fastp v0.20.0 (Chen et al. 2018), raw RNA-seq data was de novo assembled using Trinity v2.11 (Grabherr et al. 2011) with default parameters. Coding regions were identified and translated into amino acid sequences using Transdecoder v5.5.0 (https://github.com/TransDecoder/). Sequences not belonging to the target species (i.e., nonmetazoan contaminants) were filtered out using BlobTools 2 (Chen et al. 2018; Challis et al. 2020), after identification using Diamond (Buchfink et al. 2021) BLAST-P-based searches against the NCBI RefSeq protein database (https://www.ncbi.nlm.nih.gov/refseq/). Finally, only the longest isoform for each transcript was retained, representing the candidate gene's coding region. For those species with whole-genome sequences available, the predicted proteome was directly downloaded from the NCBI (https://www.ncbi.nlm.nih.gov/genome) or Uniprot (https://www.uniprot.org/proteomes) repositories. The completeness of the gene repertoire data for each species was assessed against the Metazoa set of Benchmarking Universal Single-Copy Orthologs (using BUSCO v4.1.4; Manni et al. 2021) discarding those that had a completeness score (measured as complete plus fragmented genes) lower than 80%, with only a few exceptions.
Orthology Inference Phylogenetic Tree and Molecular Dating
Orthology relationships among individual sequences for all the 117 species were inferred using OrthoFinder v2.5.1 (Emms and Kelly 2019) with default parameters. This resulted in 49,668 OGs (OGs hereafter; a proxy for gene families) with more than three sequences. From this full data set, only two outgroup species (fig. 1) were retained in downstream analyses, which resulted, after also removing low-quality sequences (see below), in 45,844 OGs (supplementary table S14, Supplementary Material online).
A topology depicting evolutionary relationships for the species in our data set was manually built based on the most recent phylogenetic studies of the clade (e.g., Kocot et al. 2019; Cunha and Giribet 2019; Anderson and Lindgren 2021). This topology was used to obtain a time-calibrated phylogenetic tree as follows. The first step consists in identifying a suitable set of orthologous genes to properly estimate branch molecular rates. To do so, Prequal v1.02 (Whelan et al. 2018) was used first to mask and filter out nonhomologous and low-quality sequence stretches from each of the OGs. As some sequences were fully filtered, the number of OGs with more than three sequences decreased (see above). Next, the masked OG sequences were aligned using MAFFT 7.4 (Katoh and Standley 2013) with automatic algorithm selection. From each of the aligned OGs, a corresponding phylogenetic tree was estimated. For OGs containing more than 500 sequences, this was accomplished using FastTree 2.1.11 (Price et al. 2010) and the LG + CAT model, as more accurate methodologies were extremely expensive computationally for alignments of this size. Trees for OGs smaller than 500 sequences were estimated with IQ-TREE 2.1 (Minh et al. 2020) under the best-fitting amino acid substitution model selected among ∼95 different models, including mixture models (estimated in IQ-TREE 1.6.12; Kalyaanamoorthy et al. 2017). To further reduce computation time, a guide tree estimated with FastTree was provided to IQ-TREE for the OGs that had mixture models as the best-fitting. Ultrafast bootstrap support using 1,000 iterations was computed for all IQTree-estimated trees. Following, PhyloPyPruner (https://gitlab.com/fethalen/phylopypruner) was employed to identify within each OG a set of truly orthologous sequences (i.e., sequences related only through speciation). This resulted in 246 OG alignments containing at least 80% of the species in the data set. These pruned OGs were further analyzed with GeneSortR (Mongiardino Koch 2021) to sort and score them based on several statistics that summarize relevant properties for their use in phylogenetic analyses (e.g., rate, discordance, saturation, etc.). The 50 genes with the highest scores (indicating less chance of being outliers and therefore minimizing systematic error in downstream analyses) were selected for the dating analysis (a number that represents a good balance between information content and computational tractability), concatenated in a supermatrix, and 10 partitions were defined based on their rate estimates.
Finally, the divergence time estimation was carried out in MCMCtree v4.8, under an autocorrelated log-normal clock and the approximate likelihood method (Reis and Yang 2011), using nine calibration points (supplementary fig. S4 and table S15, Supplementary Material online). Two parallel chains were run for 600,000 generations with a sample frequency of 10,000, a burn-in period of 400,000 generations, and checked for convergence.
Detection of Fast-Evolving Gene Families and Repertoire Reconstructions
We modeled the evolutionary dynamics of gene family size for each of the OGs using the phylogenetic “Gain-Death” (GD) model implemented in a maximum-likelihood framework in BadiRate v1.35 (Librado and Rozas 2022). In these models, gene copies in a family are gained and lost stochastically along each lineage at given gain and loss rates. If the gene gain rate is larger than the loss rate, the family tends to increase in size over time and vice versa.
More specifically, to identify OGs evolving under a different evolutionary regime in nonmarine lineages, we compared, for each OG and using a likelihood ratio test (LRT), the fit of two versions of the GD model to the OG size data (i.e., number of gene copies per species): a “global rates” model (where a single pair of gain and loss rates governs the process of gene family size evolution in the whole tree) against a “local rates” model, where rates can differ between sets of predefined branches (here between marine (background) and nonmarine [foreground] branches; figure 1). The local rates model was deemed as a significantly better explanation of the evolution of a given OG if the FDR adjusted P-value of the LRT was lower than 0.05. Then, for the OGs in which the local rates model was a better fit, the estimated net rates (gain minus loss rate) in background and foreground lineages were compared with detect those OGs that were fast-expanding/contracting (e.g., positive/negative foreground net rates and higher/lower than in the background for expanding and contracting, respectively) in the nonmarine lineages versus the marine ones. Additionally, from the best-fitting model for each OG, we obtained the maximum-likelihood reconstruction of the OG's size along the minimum number of gene gains and losses at each node. From this information, we identified expanded OGs in the lineages of interest, along full repertoire reconstructions.
Functional Annotation and Enrichment
Amino acid sequences were annotated using eggNOG-mapper v2.1.6 (Cantalapiedra et al. 2021), an orthology-based, evolutionarily informed method to transfer and assign functional information. We retained and analyzed annotations from the GO biological processes and COG databases, as well as the “preferred name” (usually the human ortholog gene name) and “description” fields. As annotations for each OG were initially composed of the pooled annotations of all the sequences contained in the OG, we obtained a concise description for each OG by retaining only the most frequent annotation. In the case of GO terms, we retained only those that were annotated to at least 10% of the annotated sequences (an empirical threshold that allowed us to remove outliers and reduce noise, while minimizing information loss). Overrepresentation tests to obtain enriched terms associated with sets of OGs were performed using ClusterProfiler (Wu et al. 2021). The background set of OGs was defined as the molluskan “pangenome” (i.e., the combination of all OGs in the data set).
To explore potential functional convergence among OGs, we developed an approach based on semantic similarity estimations and clustering that we implemented in an R package named constellatoR (https://github.com/MetazoaPhylogenomicsLab/constellatoR). Briefly, we first calculated a pairwise similarity matrix among all the concerned OGs (e.g., exclusively expanded OGs in transition lineages) based on their GO annotations using the GOSemSim package (Wu et al. 2021; Yu 2020) under the Wang semantic similarity metric (Wang et al. 2007). Next, the Affinity Propagation cluster algorithm (Frey and Dueck 2007), which does not require a desired number of clusters as input (contrary to other methods), was applied to this matrix to group OGs based on their semantic similarity (a proxy for functional overlap). Functional clusters composed of OGs from different lineages were considered as functionally convergent. Finally, using classical multidimensional scaling to reduce dimensionality, a semantic space plot was generated to represent each OG and the clustering in the functional space. We name this plot as a “constellation plot.” The ggplot2 (Wickham 2016) and ggtree packages (Yu et al. 2017) for R were used to make all plots in this manuscript.
Supplementary Material
Acknowledgments
L.A. acknowledges funding from a Juan de la Cierva-Formación fellowship (grant agreements FJC2019-042184-I funded by MCIN/AEI/10.13039/501100011033). R.F. acknowledges support from the following sources of funding: Ramón y Cajal fellowship (grant agreement no. RYC2017-22492 funded by MCIN/AEI/10.13039/501100011033 and ESF “Investing in your future’), the Agencia Estatal de Investigación (project PID2019-108824GA-I00 funded by MCIN/AEI/10.13039/501100011033), the European Research Council (this project has received funding from the European Research Council [ERC] under the European Union Horizon 2020 research and innovation program [grant agreement no. 948281]), and the Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya (AGAUR 2021-SGR00420). We also thank Centro de Supercomputación de Galicia (CESGA) for access to computer resources, and particularly Pablo Rey for his kind assistance and guidance. L.A. also thanks Pau Balart-García, Vanina Tonzo, and Federico Hoffmann for useful discussions during the development of this project.
Contributor Information
Leandro Aristide, Metazoa Phylogenomics Laboratory Biodiversity Program, Institute of Evolutionary Biology (Spanish Research Council-University Pompeu Fabra), Barcelona, Spain.
Rosa Fernández, Metazoa Phylogenomics Laboratory Biodiversity Program, Institute of Evolutionary Biology (Spanish Research Council-University Pompeu Fabra), Barcelona, Spain.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
All processed data and scripts to reproduce the analyses are deposited in the GitHub repository (https://github.com/MetazoaPhylogenomicsLab/Aristide_Fernandez_2022_mollusk_terrestrialization).
Literature Cited
- Aguilera F, McDougall C, Degnan BM. 2017. Co-option and de novo gene evolution underlie molluscan shell diversity. Mol Biol Evol. 34(4):779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat Rev Genet. 17(7):379–391. [DOI] [PubMed] [Google Scholar]
- Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. 2011. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 39(Database issue):D289–D294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson FE, Lindgren AR. 2021. Phylogenomic analyses recover a clade of large-bodied decapodiform cephalopods. Mol Phylogenet Evol. 156:107038. [DOI] [PubMed] [Google Scholar]
- Arroyo JP, Kahle KT, Gamba G. 2013. The SLC12 family of electroneutral cation-coupled chloride cotransporters. Mol Aspects Med. 34(2–3):288–298. [DOI] [PubMed] [Google Scholar]
- Barker GM. 2001. The biology of terrestrial molluscs. Wallingford, UK: CABI. [Google Scholar]
- Basse WC, et al. 2015. A sea urchin Na(+)K(+)2Cl(-) cotransporter is involved in the maintenance of calcification-relevant cytoplasmic cords in Strongylocentrotus droebachiensis Larvae. Comp Biochem Physiol Part A Mol Integr Physiol. 187(September):184–192. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, et al. 2016. Comparative genomics explains the evolutionary success of reef-forming corals. eLife 5:e13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Lenski RE, Losos JB. 2018. Contingency and determinism in evolution: replaying life's tape. Science. 362:eaam5979. [DOI] [PubMed] [Google Scholar]
- Buchfink, B, Reuter K, and Drost H-G. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 18(4): 366–368. doi: 10.1038/s41592-021-01101-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczek O, Bulaj G, Olivera BM. 2005. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci: CMLS. 62(24):3067–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambi A, Koopman M, Figdor CG. 2005. How C-type lectins detect pathogens. Cell Microbiol. 7(4):481–488. [DOI] [PubMed] [Google Scholar]
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-Mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. 38(12):5825–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R, Richards E, Rajan J, Cochrane G, Blaxter M. 2020. Blobtoolkit—interactive quality assessment of genome assemblies. G3 (Bethesda) 10(4):1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Krinsky BH, Long M. 2013. New genes as drivers of phenotypic evolution. Nat Rev Genet. 14(9):645–660. doi: 10.1038/nrg3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CY, Ko BC. 2013. NFAT5 In cellular adaptation to hypertonic stress—regulations and functional significance. J Mol Signal. 8(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Hall Matthew D.. 2018. Genetic constraints on adaptation: a theoretical primer for the genomics era. Annals of the New York Academy of Sciences. 1422(1):65–87. doi: 10.1111/nyas.2018.1422.issue-1 [DOI] [PubMed] [Google Scholar]
- Cunha TJ, Giribet G. 2019. A congruent topology for deep gastropod relationships. Proc Biol Sci/R Soc. 286(1898):20182776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CB, et al. 1999. Surfactant in the gas mantle of the snail Helix aspersa. Physiol Biochem Zool: PBZ. 72(6):691–698. [DOI] [PubMed] [Google Scholar]
- Daniels CB, Orgeig S. 2001. The comparative biology of pulmonary surfactant: past, present and future. Comp Biochem Physiol Part A Mol Integr Physiol. 129(1):9–36. [DOI] [PubMed] [Google Scholar]
- Davison A, Blaxter M. 2005. Ancient origin of glycosyl hydrolase family 9 cellulase genes. Mol Biol Evol. 22(5):1273–1284. [DOI] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW. 2009. The life and death of gene families. BioEssays News Rev Mol Cell Dev Biol. 31(1):29–39. [DOI] [PubMed] [Google Scholar]
- Denisov IG, Makris TM, Sligar SG, Schlichting I. 2005. Structure and chemistry of cytochrome P450. Chem Rev. 105(6):2253–2277. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2015. Orthofinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DH. 2015. Novelty and innovation in the history of life. Curr Biol: CB. 25(19):R930–R940. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Phylogenies and the comparative method. Am Nat. 125(1):1–15. [Google Scholar]
- Fernández R, et al. 2022. MATEdb, a data repository of high-quality metazoan transcriptome assemblies to accelerate phylogenomic studies. Peer Commun J. 2:e58. [Google Scholar]
- Foulquier F, et al. 2012. TMEM165 deficiency causes a congenital disorder of glycosylation. Am J Hum Genet. 91(1):15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey BJ, Dueck D. 2007. Clustering by passing messages between data points. Science 315(5814):972–976. [DOI] [PubMed] [Google Scholar]
- Giglio ML, Ituarte S, Pasquevich MY, Heras H. 2016. The eggs of the apple snail Pomacea maculata are defended by indigestible polysaccharides and toxic proteins. Can J Zool. 94(11):777–785. [Google Scholar]
- Goldstone JV, et al. 2006. The chemical defensome: environmental sensing and response genes in the strongylocentrotus Purpuratus genome. Dev Biol. 300(1):366–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B. 2009. The causes of repeated genetic evolution. Dev Biol. 332(1):36–47. [DOI] [PubMed] [Google Scholar]
- Gorson J, et al. 2015. Molecular diversity and gene evolution of the venom arsenal of Terebridae predatory marine snails. Genome Biol Evol. 7(6):1761–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Vrba ES. 1982. Exaptation—a missing term in the science of form. Paleobiology 8(1):4–15. [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf DL, Cummings KS. 2007. Review of the systematics and global diversity of freshwater mussel species (Bivalvia: Unionoida). J Molluscan Stud. 73(4):291–314. [Google Scholar]
- Hallinan NM, Lindberg DR. 2011. Comparative analysis of chromosome counts infers three paleopolyploidies in the Mollusca. Genome Biol Evol. 3:1150–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 1994. The eIF-2 alpha kinases: regulators of protein synthesis in starvation and stress. Semin Cell Biol. 5(6):417–426. [DOI] [PubMed] [Google Scholar]
- Hu MY, et al. 2018. A SLC4 family bicarbonate transporter is critical for intracellular pH regulation and biomineralization in sea urchin embryos. eLife 7:e36600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X-D, Wei G-J, Zhang H, He M-X. 2015. Nuclear factor of activated T cells (NFAT) in pearl oyster Pinctada fucata: molecular cloning and functional characterization. Fish Shellfish Immunol. 42(1):108–113. [DOI] [PubMed] [Google Scholar]
- Jacob F. 1977. Evolution and tinkering. Science 196(4295):1161–1166. [DOI] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser P, Borge-Renberg K, Hultmark D. 2007. The Drosophila NFAT homolog is involved in salt stress tolerance. Insect Biochem Mol Biol. 37(4):356–362. [DOI] [PubMed] [Google Scholar]
- Kocot KM, Todt C, Mikkelsen NT, Halanych KM. 2019. Phylogenomics of aplacophora (Mollusca, Aculifera) and a Solenogaster without a foot. Proc Biol Sci/R Soc. 286(1902):20190115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh A, Walter P. 2012. Structural basis of the unfolded protein response. Annu Rev Cell Dev Biol. 28:251–277. [DOI] [PubMed] [Google Scholar]
- Krug PJ, et al. 2022. Phylogenomic resolution of the root of Panpulmonata, a hyperdiverse radiation of gastropods: new insight into the evolution of air breathing. Proc Biol Sci/R Soc. 289(1972):20211855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse E, Uehlein N, Kaldenhoff R. 2006. The aquaporins. Genome Biol. 7(2):206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, et al. 2010. The sodium bicarbonate cotransporter (NBCe1) is essential for normal development of mouse dentition. J Biol Chem. 285(32):24432–24438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2022. Reconstructing gene gains and losses with BadiRate. Methods Mol Biol. 2569:213–232. [DOI] [PubMed] [Google Scholar]
- Little C, Honorary Research Associate Colin Little . 1983. The colonisation of land: origins and adaptations of terrestrial animals. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Liu C, et al. 2018. The genome of the golden apple snail Pomacea canaliculata provides insight into stress tolerance and invasive adaptation. GigaScience 7(9):giy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, et al. 2021. Giant African snail genomes provide insights into molluscan whole-genome duplication and aquatic-terrestrial transition. Mol Ecol Resour. 21(2):478–494. [DOI] [PubMed] [Google Scholar]
- Livingstone DR, de Zwaan A. 1983. Metabolic biochemistry and molecular biomechanics. Amsterdam, Netherlands: Elsevier. p. 177–242. [Google Scholar]
- Lorgen M, Jorgensen EH, Jordan WC, Martin SAM, Hazlerigg DG. 2017. NFAT5 genes are part of the osmotic regulatory system in Atlantic salmon (Salmo salar). Mar Genomics. 31:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos JB. 2011. Convergence, adaptation, and constraint. Evolution. 65(7):1827–1840. [DOI] [PubMed] [Google Scholar]
- Luckenbach T, Epel D. 2008. ABCB- and ABCC-type transporters confer multixenobiotic resistance and form an environment-tissue barrier in bivalve gills. Am J Physiol Regul Integr Comp Physiol. 294(6):R1919–R1929. [DOI] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 38(10):4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Redondo GI, et al. 2023. Parallel duplication and loss of aquaporin-coding genes during the ‘out of the sea’ transition as potential key drivers of animal terrestrialization. Mol Ecol. 32(8):2022–2040. [DOI] [PubMed] [Google Scholar]
- McDougall C, Aguilera F, Degnan BM. 2013. Rapid evolution of pearl oyster shell matrix proteins with repetitive, low-complexity domains. J R Soc Interface/R Soc. 10(82):20130041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane A, et al. 2021. Mucus. Curr Biol: CB. 31(15):R938–R945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, et al. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongiardino Koch N. 2021. Phylogenomic subsampling and the search for phylogenetically reliable loci. Mol Biol Evol. 38(9):4025–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumoff DG. 2011. Hierarchical classification of glycoside hydrolases. Biochem Biokhimiia. 76(6):622–635. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1985. Dispensable genes. Trends Genet. 1:160–164. [Google Scholar]
- Pizzagalli MD, Bensimon A, Superti-Furga G. 2021. A guide to plasma membrane solute carrier proteins. FEBS J. 288(9):2784–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder W, Lindberg DR. 2008. Phylogeny and evolution of the Mollusca. Berkeley, CA: University of California Press. [Google Scholar]
- Popper ZA, et al. 2011. Evolution and diversity of plant cell walls: from algae to flowering plants. Annu Rev Plant Biol. 62:567–590. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. Fasttree 2–approximately maximum-likelihood trees for large alignments. PloS One. 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis MD, Yang Z. 2011. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol. 28(7):2161–2172. [DOI] [PubMed] [Google Scholar]
- Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. 2012. Glycogen and its metabolism: some new developments and old themes. Biochem J. 441(3):763–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero PE, Pfenninger M, Kano Y, Klussmann-Kolb A. 2016. Molecular phylogeny of the Ellobiidae (Gastropoda: Panpulmonata) supports independent terrestrial invasions. Mol Phylogenet Evol. 97:43–54. [DOI] [PubMed] [Google Scholar]
- Romero PE, Weigand AM, Pfenninger M. 2016. Positive selection on panpulmonate mitogenomes provide new clues on adaptations to terrestrial life. BMC Evol Biol. 16(1):164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum EB, Parent CE, Brandt EE. 2014. The molecular basis of phenotypic convergence. Annu Rev Ecol Evol Syst. 45:203–226. [Google Scholar]
- Salzet M, Deloffre L, Breton C, Vieau D, Schoofs L. 2001. The angiotensin system elements in invertebrates. Brain Res Brain Res Rev. 36(1):35–45. [DOI] [PubMed] [Google Scholar]
- Selden P. 2012. Terrestrialisation (Precambrian–Devonian). eLS. 2:1–6. [Google Scholar]
- Shear WA. 1991. The early development of terrestrial ecosystems. Nature. 351:283–289. [Google Scholar]
- Sokolova IM, Frederich M, Bagwe R, Lannig G, Sukhotin AA. 2012. Energy homeostasis as an integrative tool for assessing limits of environmental stress tolerance in aquatic invertebrates. Mar Environ Res. 79:1–15. [DOI] [PubMed] [Google Scholar]
- Sorensen JS, Denise Dearing M. 2006. Efflux transporters as a novel herbivore countermechanism to plant chemical defenses. J Chem Ecol. 32(6):1181–1196. [DOI] [PubMed] [Google Scholar]
- Sørensen JG, Kristensen TN, Loeschcke V. 2003. The evolutionary and ecological role of heat shock proteins. Ecol Lett. 6(11):1025–1037. [Google Scholar]
- Stayton CT. 2015. What does convergent evolution mean? The interpretation of convergence and its implications in the search for limits to evolution. Interface Focus. 5(6):20150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange RC, Spiteri MA, Ramachandran S, Fryer AA. 2001. Glutathione S-transferase family of enzymes. Mutat Res. 482(1–2):21–26. [DOI] [PubMed] [Google Scholar]
- Strong EE, Gargominy O, Ponder WF, Bouchet P. 2007. Global diversity of gastropods (Gastropoda; Mollusca) in freshwater. Dev Hydrobiol. 198:149–166. [Google Scholar]
- Sun J, et al. 2017. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat Ecol & Evol. 1(5):121. [DOI] [PubMed] [Google Scholar]
- Sun J, et al. 2019. Signatures of divergence, invasiveness, and terrestrialization revealed by four apple snail genomes. Mol Biol Evol. 36(7):1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28(1):33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeij GJ, Watson-Zink VM. 2022. Terrestrialization in gastropods: lineages, ecological constraints and comparisons with other animals. Biol J Linn Soc. 136(3):393–404. [Google Scholar]
- Wang J, et al. 2020. Genomic and transcriptomic landscapes and evolutionary dynamics of Molluscan glycoside hydrolase families with implications for algae-feeding biology. Comput Struct Biotechnol J. 18:2744–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, et al. 2021. African lungfish genome sheds light on the vertebrate water-to-land transition. Cell 184(5):1362–76.e18. [DOI] [PubMed] [Google Scholar]
- Wang JZ, Du Z, Payattakool R, Yu PS, Chen C-F. 2007. A new method to measure the semantic similarity of GO terms. Bioinformatics 23(10):1274–1281. [DOI] [PubMed] [Google Scholar]
- Whelan S, Irisarri I, Burki F. 2018. PREQUAL: detecting non-homologous characters in sets of unaligned homologous sequences. Bioinformatics 34(22):3929–3930. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2016. Ggplot2: elegant graphics for data analysis. New York, US: Springer. [Google Scholar]
- Wood ZA, Schröder E, Robin Harris J, Poole LB. 2003. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 28(1):32–40. [DOI] [PubMed] [Google Scholar]
- WoRMS Editorial Board . 2022. World Register of Marine Species. Available fromhttps://www.marinespecies.orgat VLIZ. Accessed Yyyy-Mm-Dd.” VLIZ. 10.14284/170. [DOI]
- Wright BJ, Bickham-Wright U, Yoshino TP, Jackson MB. 2017. H+ channels in embryonic Biomphalaria glabrata cell membranes: putative roles in snail host-schistosome interactions. PLoS Negl Trop Dis. 11(3):e0005467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, et al. 2021. Clusterprofiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2(3):100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, et al. 2018. Molecular cloning and characterization of two genes encoding peroxiredoxins from freshwater bivalve Anodonta woodiana: antioxidative effect and immune defense. Fish Shellfish Immunol. 82:476–491. [DOI] [PubMed] [Google Scholar]
- Yarra T, Blaxter M, Clark MS. 2021. A bivalve biomineralization toolbox. Mol Biol Evol. 38(9):4043–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. 2020. Gene ontology semantic similarity analysis using GOSemSim. Methods Mol Biol. 2117:207–215. [DOI] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. Ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol/Br Ecol Soc. 8(1):28–36. [Google Scholar]
- Zhang G, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490(7418):49–54. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. 2015. Massive expansion and functional divergence of innate immune genes in a protostome. Sci Rep. 5:8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. 2021. The genome of Nautilus pompilius illuminates eye evolution and biomineralization. Nat Ecol Evol. 5(7):927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li Q, Yu H, Liu S, Kong L. 2021. Shell biosynthesis and pigmentation as revealed by the expression of tyrosinase and tyrosinase-like protein genes in pacific oyster (Crassostrea gigas) with different shell colors. Mar Biotechnol. 23(5):777–789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All processed data and scripts to reproduce the analyses are deposited in the GitHub repository (https://github.com/MetazoaPhylogenomicsLab/Aristide_Fernandez_2022_mollusk_terrestrialization).