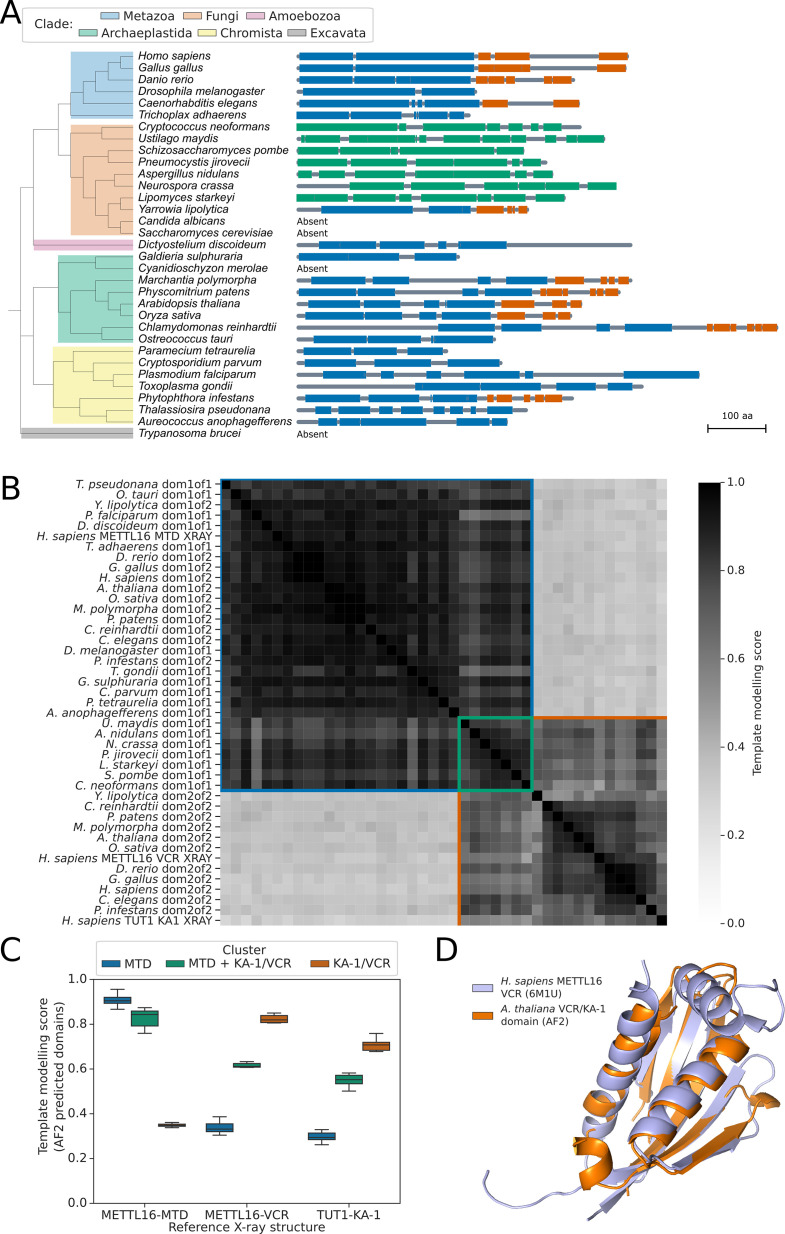
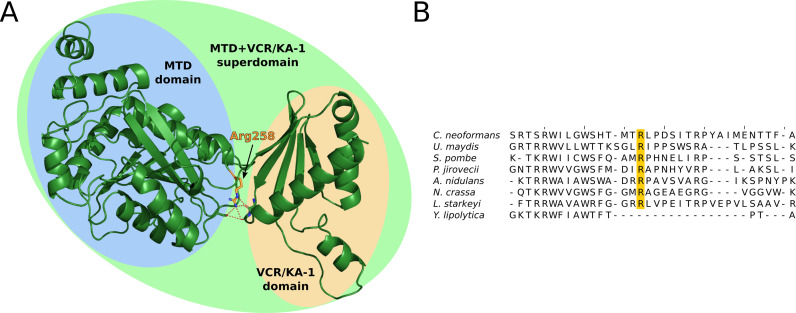
Figure 1. METTL16 is widely conserved across eukaryotes.
(A) Phylogenetic tree showing the presence and absence of a METTL16 ortholog in 33 eukaryotic species. Linear protein structures with globular domains identified from Alphafold2 models are shown on the right of the tree. Domains are colored by cluster: MTD domains in blue, MTD +VCR/KA-1 domains in green, and VCR/KA-1 domains in orange. Likely loop/disordered regions with low confidence predictions are shown as grey lines. (B) Distance matrix heatmap showing the pairwise TMscore of segmented domains from the Alphafold2 predictions of 29 METTL16 orthologs. The X-ray structures of human METTL16 MTD and VCR, and TUT1 KA-1 are included as positive controls. Domains are grouped into clusters (diagonal boxes) using the same color scheme as in (A). (C) Boxplot showing TMscores of segmented domains from Alphafold2 predictions of 28 METTL16 orthologs superimposed onto experimentally determined structures of human METTL16 MTD and VCR, and TUT1 KA-1. (D) Superimposition of the VCR/KA-1 domain of Arabidopsis FIO1 predicted by Alphafold2 onto the X-ray structure of the human METTL16 VCR.