Fig. 2.
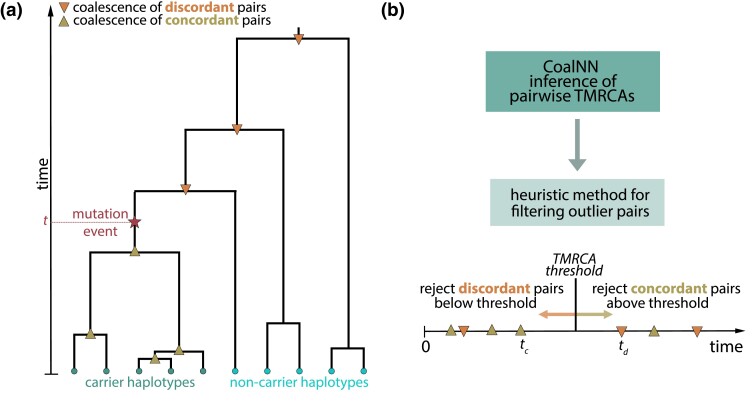
Procedure for dating genomic variants. (a) At a given genomic site, individuals are connected through underlying genealogical relationships. We aim to infer the time t at which a mutation arose (denoted by the star) and resulted in carrier haplotypes and noncarrier haplotypes. (b) When dating variants, CoalNN first infers TMRCAs across all concordant (two carriers) and discordant (one carrier and one noncarrier) pairs of haplotypes and then rejects outlier pairs using the heuristic approach developed in Albers and McVean (2020). The TMRCA rejection threshold is computed by minimizing the total number of rejected pairs. The predicted age estimate for the variant is obtained by averaging the maximum coalescence time across concordant pairs and the minimum coalescence time across discordant pairs after filtering.