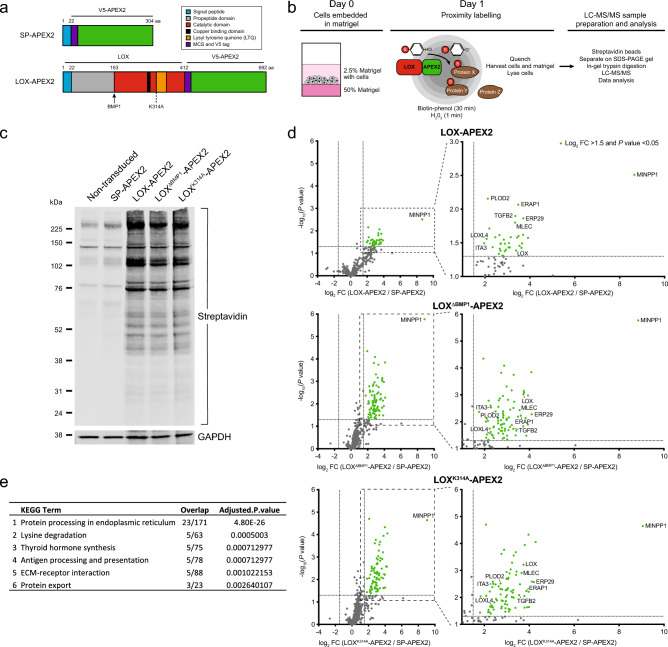
Figure 1.
APEX2-based proximity labelling identifies LOX interacting proteins. (a) Schematic diagram of SP-APEX2 and LOX-APEX2 constructs, with domain structures, and positions of the BMP1 cleavage site and K314A mutation. Numbers above: amino acids. (b) Schematic diagram of APEX2-labelling experimental approach for LOX interacting proteins. (c) Western blot for biotinylated proteins revealed by streptavidin binding to immobilised proteins in lysates from MDA-MB-231 cells stably expressing SP-APEX2 or the indicated LOX-APEX2 variants. GAPDH: loading control. Representative image of 3 independent experiments. (d) Volcano plots showing proteins interacting with the indicated LOX-APEX2 variants compared to SP-APEX2 controls in MDA-MB-231 cells. Expanded regions highlight the most significant interactors. Green dots: log2 abundance fold change (FC) > 1.5, P < 0.05). Data are from three independent biological replicates. (e) KEGG pathway analysis for the top five significantly enriched pathways for LOX interacting proteins.