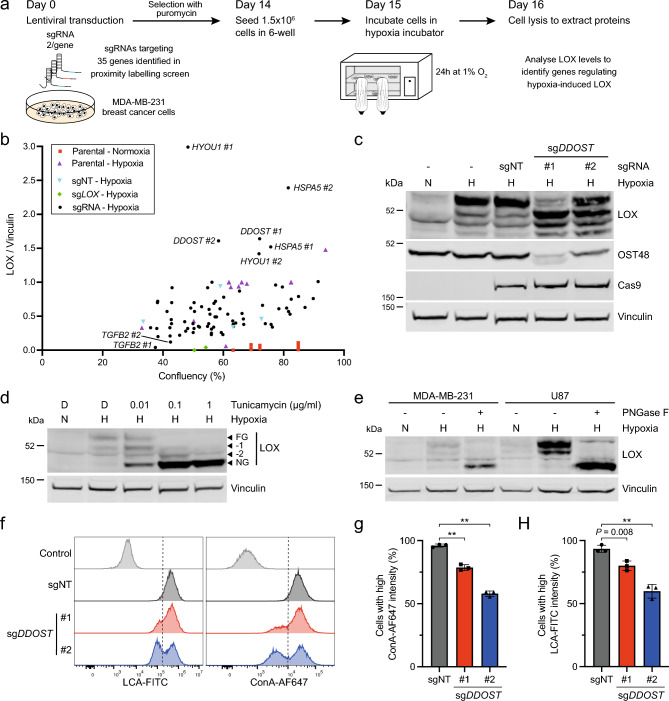
Figure 2.
DDOST depletion suppresses LOX and cell surface N-glycosylation. (a) Schematic diagram of genome editing experimental approach to test how depletion of LOX-interactors affect LOX expression in hypoxic MDA-MB-231 cells. Cells were transduced with LentiCRISPRv2-sgRNA vectors targeting 35 genes identified in Fig. 1d (LOX-APEX2 interactors), re-seeded 14 days later, transferred to 1% O2 the next day and harvested for western blot analysis 24 h later. (b) Graph showing LOX expression (fold change compared to vinculin) as a function of cell confluency (%) in MDA-MB-231 cells after CRISPR/Cas9 gene editing for 35 genes selected from Fig. 1d (LOX-APEX2 interactors). Two optimal sgRNAs per gene were used alongside three independent non-targeting controls (sgNT; cyan) and two sgLOX (green) controls. Cell confluency was determined by IncuCyte imaging 15 days after transduction, then the cells were placed in 1% O2 for 24 h (except the MDA-MB-231 parental control; red) and harvested for LOX quantification by western blot. (c) Immunoblot for LOX, OST48, Cas9 and vinculin (loading control) in MDA-MB-231 cells after CRISPR/Cas9 gene editing with sgDDOST (#1, #2) or sgNT (control) for 15 days and then incubated in normoxia (N) or hypoxia (H; 1% O2) for 24 h. Image representative of 3 independent experiments. (d) Western blot for LOX and vinculin (loading control) in MDA-MB-231 cells treated with the indicated concentrations of tunicamycin or DMSO (D) control and incubated in normoxia (N) or hypoxia (H; 1% O2) for 24 h. FG: fully N-glycosylated; NG: non-glycosylated; −1, −2 incomplete N-glycosylation (intermediate). Image representative of 3 independent experiments. (e) Western blot for LOX and vinculin (loading control) in PNGase F- (+) or vehicle-treated (-) lysates from MDA-MB-231 and U87 cells incubated in normoxia (N) or hypoxia (H; 1% O2) for 24 h. Image representative of 3 independent experiments. (f) Histograms showing LCA and ConA binding in U87 cells 14 days after CRISPR/Cas9 gene editing with sgDDOST (#1, #2) or sgNT (control). The control shows unstained cells and data are representative of 3 independent experiments. (g, h) Quantification of ConA (g) and LCA (h) binding in U87 cells 14 days after CRISPR/Cas9 gene editing with sgDDOST (#1, #2) or sgNT (control). Data are mean ± s.d. of 3 independent experiments. ** P < 0.001.