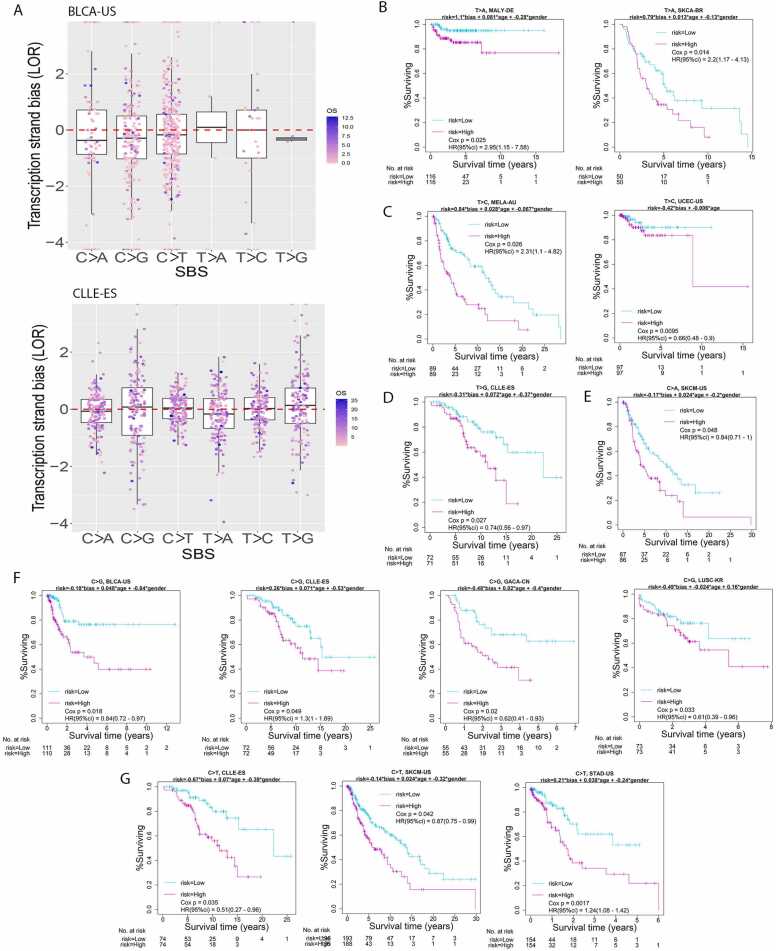
Fig. 4.
Quantification of individual-level transcription strand bias and its association with cancer survival. A. Patients in one were each assessed for their prominence of transcription strand bias with respect to a mutation category. Shown were two example cohorts: BLCA-US (top) and CLLE-ES (bottom). B. MALY-DE and SKCA-BR showed prognosis significance for T > A transcription strand bias. C. MELA-AU and UCEC-US showed prognosis significance for T > C transcription strand bias. D. CLLE-DE showed prognosis significance for T > G transcription strand bias. E. SKCM-US showed prognosis significance for C>A transcription strand bias. F. BLCA-US, CLLE-ES, GACA-CN, and LUSC-KR showed prognosis significance for C>G transcription strand bias. G. CLLE-ES, SKCM-US, and STAD-US showed prognosis significance for C>T transcription strand bias.