Summary
Inhibitory neurons are essential for orchestrating and structuring behavior. We use one of the best studied behaviors in Drosophila, male courtship, to analyze how inhibitory, GABAergic neurons shape the different steps of this multifaceted motor sequence. RNAi-mediated knockdown of the GABA-producing enzyme GAD1 and the ionotropic receptor Rdl in sex specific, fruitless expressing neurons in the ventral nerve cord causes uncoordinated and futile copulation attempts, defects in wing extension choice and severe alterations of courtship song. Altered song of GABA depleted males fails to stimulate female receptivity, but rescue of song patterning alone is not sufficient to rescue male mating success. Knockdown of GAD1 and Rdl in male brain circuits abolishes courtship conditioning. We characterize the around 220 neurons coexpressing GAD1 and Fruitless in the Drosophila male nervous system and propose inhibitory circuit motifs underlying key features of courtship behavior based on the observed phenotypes.
Subject areas: Biological sciences, Neuroscience, Behavioral neuroscience, Molecular neuroscience
Graphical abstract

Highlights
-
•
GABAergic inhibition in fruitless+ neurons shapes Drosophila male courtship
-
•
Inhibition gates copulation attempts
-
•
Inhibition determines unilateral wing posture during courtship singing
-
•
GAD1+ and Rdl+ inhibitory motifs in the ventral nerve cord pattern courtship song
Biological sciences; Neuroscience; Behavioral neuroscience; Molecular neuroscience
Introduction
Neuronal inhibition is a universal feature of nervous systems. Fast neuronal inhibition is accomplished by GABA, a neurotransmitter conserved across vertebrates and invertebrates that is already present in the simple nerve nets of cnidaria.1,2,3 GABAergic inhibition in the nervous system is important for stabilizing and shaping network activity and counterbalancing excitation, allowing for efficient coding and preventing runaway excitation and epileptic seizure.4,5,6,7,8 Additional to this global and general role, inhibitory connections shape circuit motifs that enable computations performed in many different pathways across many different species.9,10 Such circuit motifs support effective sensory processing, and adaptive behavioral choice in response to external stimuli and internal state.11,12,13,14 For example, inhibitory interneuron networks enable male moths to track female pheromone plumes, crickets to process the temporal structure of the intraspecific acoustic calls and honeybees to decode the waggle dance vibrational pattern of their nestmates.15 In the Drosophila larvae nervous system, inhibitory circuit motifs based on reciprocal inhibition, lateral disinhibition and feedback disinhibition allow the animal to respond to mechanical stimuli with two distinct motor programs, either head turning or head retracting.16
Here, we use Drosophila male courtship behavior to address the question how neuronal inhibition shapes a complex behavioral sequence. Courtship consists of multiple coordinated and ordered steps relying on multimodal sensory integration and the generation of specific, precisely timed motor patterns. Characteristic courtship displays are the male tracking and following the female, tapping her abdomen with the foreleg to sample pheromones and vibrating one extended wing. The wing vibrations produce a highly structured acoustic signal, the courtship song that stimulates the female’s receptivity and promotes her acceptance of copulation.17,18,19 When the female slows down, the male attempts copulation by probing the female genitalia with his proboscis, bending his abdomen and bringing his genitalia in apposition with the female genitalia. If the female opens her vaginal plates (hypogynia), copulation can occur.20,21
The neuronal circuits underlying male fly courtship are among the best-studied model circuits for behavior in the Drosophila nervous system.17,22,23,24 Most, if not all key neuronal components for male courtship express the male-specific transcription factor FruitlessM (FruM). FruM is present in around 2% cells of the adult nervous system.25,26 Among them are neurons controlling following behavior,27 singing28 and copulation attempts.29 Several studies have surveyed the distribution, anatomy, and interconnection of FruM expressing (fru+) neurons.25,26,30,31,32,33,34 However, expression of neuronal transmitter types, including GABA, in the whole FruM positive cell population, as well as their contribution to circuit function has to our knowledge not been studied in detail so far. GABAergic transmission is widespread in the Drosophila nervous system, with the GABA-producing enzyme glutamic acid decarboxylase (GAD1), the ionotropic GABA receptor Rdl and the vesicular GABA transporter (vGAT) distributed in all major neuropil areas of the brain and ventral nerve cord.35,36,37,38,39 Single-cell transcriptomics analysis suggests that around 25% of Drosophila central brain neurons40 and around 10% of the entire brain including optic lobes41 are GABAergic. In the central nerve cord, around 38% of all neurons express GAD1, the marker enzyme for GABAergic neurons.42 In these datasets, visualized by the SCope tool, co-expression of GAD1 and fruitless is obvious in numerous neurons.41
Previously, few GABAergic fruitless expressing (fru+) neurons have been investigated in detail in their role for courtship behavior. The GABAergic brain neuronal class mAL, for example, regulates the processing of gustatory information that informs the courting male of the sex of another fly. mAL provides inhibitory input to central courtship promoting P1 neurons. mAL is activated by leg gustatory neurons that sense male pheromones with courtship suppressing effect. It thus functions to prevent male-male courtship. Interestingly, mAL is also activated by different subsets of leg gustatory neurons that sense courtship-promoting female pheromones. Since the sensory neurons responsive to female pheromones activate both GABAergic mAL and the interneuron vAB3 that excites P1, its second function is probably to exert gain control in the response of P1 to courtship stimulating female pheromones.43,44
A more comprehensive view of how GABAergic inhibition shapes the entire behavioral sequence of male courtship, however, is missing. Moreover, it has never been addressed how GABAergic neurons contribute to generating the wing song motor pattern, a central element of male courtship whose correct execution strongly affects copulation success.
Here, we used RNAi-mediated knockdown of GAD1 or Rdl expression in fru+ neurons and asked how GABAergic signaling in the male-specific circuit affected courtship. We further provide an anatomical description of all GAD1+ and fru+ neurons in brain and ventral nerve cord. We test how brain versus ventral nerve cord populations contribute to the various behavioral phenotypes overserved after the disruption of GABAergic signaling. By focusing on courtship song patterning, we analyze with recording and playback experiment the causal relationship between disrupted song patterns and copulation success.
We find that the depletion of GAD1 and Rdl in fru+ neurons leads to very similar specific defects in the hierarchical organization and motor coordination of male behavior, preventing the animals from achieving copulation. Copulation is frequently attempted, but not in the appropriate proximity and orientation to the female. Males sing with both wings. Upon closer inspection of the acoustic signals generated by wing vibration, we find that knockdown of GAD1 or Rdl leads to severe alterations of the courtship song pattern. The sine song mode is strongly reduced and the pulse song mode is altered in pulse structure and spacing. This strongly altered song fails to stimulate the receptivity of wild type females. Wild-type song, however, cannot rescue the copulation defect of knockdown males, indicating that additionally to song, defects in copulation behavior prevents them from mating. Apart from the defects in the execution of the different courtship steps, GAD1 and Rdl depleted males are unable to reduce courtship after negative conditioning with unreceptive females. All observed singing and song patterning phenotypes result from the depletion of GABAergic signaling in the ventral nerve cord. In contrast, the manipulation of GABAergic neurons in the brain affects courtship conditioning.
Our study illustrates that GABAergic fru+ neurons that inhibit Rdl+ fru+ neurons tune many aspects of courtship behavior. We locate the circuit motifs for unilateral wing usage, sine song, pulse length, and pulse spacing, as well as the coordination of intromission in the ventral nerve cord. In the future, it will be interesting to identify the cellular identity and the connectivity of these motifs.
Results
GABAergic signaling in fruitless neurons is required for the motor coordination of male courtship behavior
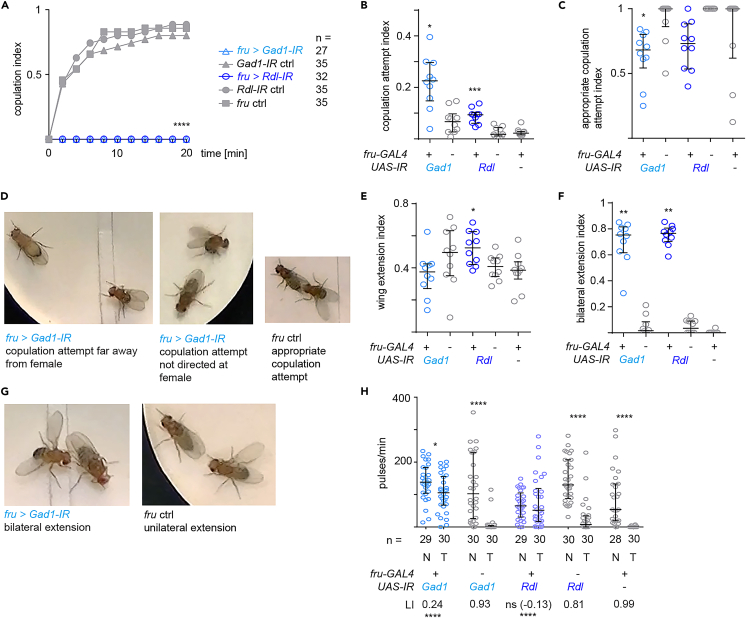
To assess the role of GABAergic signaling in male courtship behavior, we focused on the approx. 2% of adult male neurons expressing the sex determination factor FruM and analyzed males that had either the GABA-producing enzyme GAD1 or the ionotropic GABA receptor Rdl depleted in all neurons expressing the fruitless-GAL4 (fru-GAL4) driver (fru+ neurons) by RNAi mediated knockdown. In contrast to controls, knockdown males did not achieve copulation when paired with wild type virgin females for 20 min (Figure 1A). This was not due to low courtship intensity since knockdown males attempted copulation more often than controls (Figure 1B). When we scrutinized the observed copulation attempts, however, we saw that knockdown males had a lower proportion of appropriate copulation attempts, i.e., attempts that were directed at the female abdomen and performed in the close vicinity of the female (Figures 1C and 1D). Next, we asked if another central courtship element, male wing song, was affected by the depletion of GABAergic signaling. Knockdown males extended their wings as often or, in the case of Rdl knockdown, more often than control males (Figure 1E). Most of the knockdown male wing extensions differed markedly from controls in such that the male extended two wings instead of one (Figures 1F and 1G). Next, we wondered if knockdown males would abstain from singing when control males naturally do so, namely after courtship conditioning. Control males sing only very little or not at all after being trained with a mated, unreceptive female for 1 h. Males depleted of Rdl sang the same amount of pulse song with or without training. Males depleted of GAD1 showed a slight reduction of pulse song. Their learning index, however, was significantly lower than the learning indices of control flies (Figure 1H).
Figure 1.
Depletion of GABAergic inhibition affects the motor coordination of male courtship behavior
(A) Copulation index of male flies upon RNAi-mediated knockdown of GAD1 or Rdl in fruitless neurons and respective genetic controls. ∗∗∗∗p < 0.0001, Fisher exact text. n indicates number of flies tested.
(B) Copulation attempt index (fraction of video frames in which the male attempted copulation) for knockdown and control males.
(C) Appropriate copulation attempt index (fraction of copulation attempts which were directed at and in reach of the female) for knockdown and control males.
(D) Examples of inappropriate copulation attempts of a GAD1 depleted male and an appropriate copulation attempt of control male.
(E) Wing extension index (fraction of video frames in which the male extended a wing) for knockdown and control males.
(F) Bilateral wing extension index (fraction of wing extensions which were bilateral) for knockdown and control males.
(G) Example of bilateral wing extension of a GAD1 depleted male and a unilateral extension of control male.
(H) The effect of courtship conditioning in knockdown and control males. For each genotype the amount of song (in pulses/min) toward a mated wild type female is shown for naive (N) males and males trained (T) with an unreceptive mated wild type female. ∗p = 0.02, ∗∗∗∗p < 0.0001, Mann-Whitney test. The learning index of each genotype (LI) is given under the graph in the cases were significant learning occurred. The LI of the two knockdown genotypes was compared to the LIs of the two respective control genotypes by a permutation test, ∗∗∗∗p < 0.0001 indicates significant difference between LI of knockdown genotypes and the LIs of both controls. n indicates number of flies tested. (A–G) All experiments are performed with wild type virgin females. (B, C, E, F) Each knockdown genotype is compared to its corresponding UAS- IR control and the fru ctrl, ∗p < 0.05, ∗∗p < 0.005, Kruskal-Wallis test with Dunn’s multiple comparison, n = 10 males per genotype. In all scatterplots, each data point represents one fly, error bars indicate median with interquartile range.
We conclude that males depleted of GABAergic inhibition in fru+ neurons do not copulate within normal time spans, fail to suppress inappropriate courtship attempts, extend two wings during singing and show no or strongly diminished reduction of singing after courtship conditioning.
Despite defects in courtship and failure to quickly mate in observation chambers, males depleted for GAD1 in fru+ neurons were fully fertile when left for 3 days with virgin females in food vials. Males depleted for Rdl in fru+ neurons were fertile with a moderately decreased fertility of 67% as compared to 100% in control flies (Figure S1A). We therefore observed the mating behavior of knockdown males on food in the presence of 3 virgin females, conditions which have been reported to increase mating rates.45 In this experimental set up, the copulation index of knockdown males did not differ from controls and all or nearly all males achieved copulation within 12 h, although it took them significantly longer to do so (Figure S1B). Sperm transfer, a prerequisite for a fertile mating, takes place around 8 min after the initiation of genital coupling.46 We therefore only considered events of genital coupling that lasted for more than 8 min potentially fertile, full copulations. 76% of males depleted for GAD1 in fru+ neurons showed at least one copulation shorter than 8 min before they achieved a full copulation, a phenotype not seen in control males or males depleted for Rdl (Figure S1C). When omitting short copulations from evaluation, the copulation duration of males depleted for GAD1 in fru+ neurons was not significantly different from control males. In contrast, males depleted for Rdl in fru+ neurons copulated longer than control males (Figure S1D).
In summary, GABAergic inhibition in fru+ neurons is not only required in males to achieve copulation without delay, but also for correct copulation behavior. Males depleted for GAD1 in fru+ neurons show a high incidence of very short copulations, whereas males depleted for Rdl in fru+ neurons copulate longer and have a decreased fertility.
Patterning of courtship song depends on GABAergic signaling
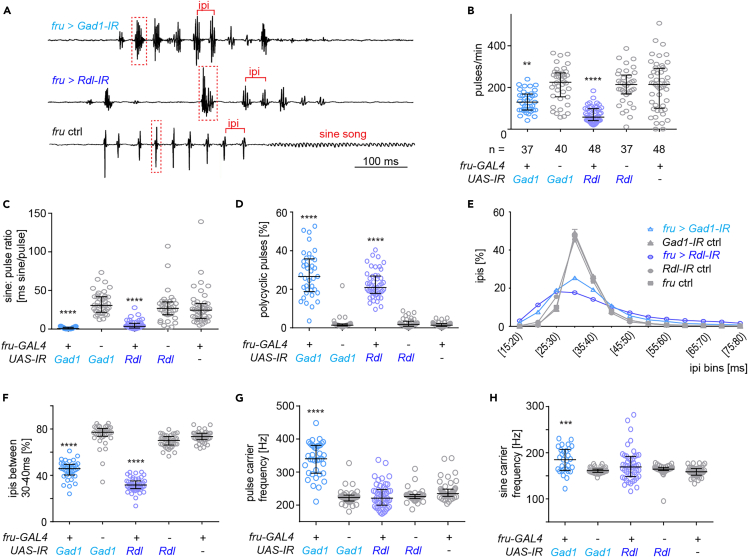
The postural defect during singing led us to the question if the depletion of GABAergic signaling in fru+ neurons also affected the patterning of the acoustic properties of courtship song. Courtship song consists of wing pulse trains (pulse song) and lower volume continuous wing oscillations (sine song). Characteristic and species-specific features of pulse song is the timing of inter pulse intervals (ipis) and the length of an individual pulse (Figure 2A). Knockdown males reliably sang pulse song, albeit a lower amount than control males (Figure 2B). As immediately apparent from sample oscillograms, song of males depleted for either GAD1 or Rdl in fru+ neurons had marked and similar changes compared to the song of control males (Figure 2A). Sine song was strongly reduced (Figure 2C) and pulse song was polycyclic, i.e., individual pulses increased in length and the number of underlying wing strokes (cycles) (Figures 2A and 2D). While the control pulse song is characterized by a high percentage of pulses which are spaced at 30–35 ms, the distribution of ipis of knockdown flies was less defined and broader, with less ipis in the 30–40 ms range (Figures 2E and 2F). After the knockdown of GAD1, but not Rdl, the carrier frequency of pulse and sine song increased (Figures 2G and 2H). To confirm that these phenotypes resulted specifically from the depletion of GABAergic signaling, we repeated knockdown experiments with independent RNAi lines against GAD1 and Rdl, as well as an RNAi line against the vesicular GABA transporter vGAT. We reasoned that knockdown of the latter, which is essential for packaging GABA into synaptic vesicles, would lead to similar phenotypes than interference with GABA synthesis. Males depleted for GABAergic signaling with these alternative RNAi lines showed very similar song defects to the ones described above (Figure S2). We conclude that GABAergic, fru+ neurons, and fru+ neurons expressing Rdl are required for the correct patterning of courtship song.
Figure 2.
GABAergic signaling in fruitless neurons is required for courtship song patterning
(A) Examples of oscillograms of courtship song from knockdown males and a control male. In the pulse trains, ipis are marked with red brackets. Individual pulses are shown inside dashed red rectangles. Note the presence of sine song in control oscillogram and its lack in the knockdown genotypes.
(B) Amount of courtship song (in pulses/min) displayed toward a virgin wild type female of male flies upon RNAi-mediated knockdown of GAD1 or Rdl in fruitless neurons and respective genetic controls. n indicates number of flies tested.
(C) Amount of sine song (sine: pulse ratio) for knockdown and control males.
(D) Amount of polycyclic pulses (% of pulses with more than 2 cycles) in the song of knockdown and control males.
(E) Distribution of inter pulse intervals (ipis) in the song of knockdown and control males. For each genotype, the mean percentage of ipis in the corresponding bin among flies and the standard error of the mean is plotted.
(F) Percentage of inter pulse intervals (ipis) within the 30–40 ms range in the song of knockdown and control males.
(G) Median pulse song carrier frequency for knockdown and control males.
(H) Median sine song carrier frequency for knockdown and control males. (A, C, D, F–H) Each knockdown genotype is compared to its corresponding UAS- IR control and the fru ctrl, ∗p = 0.017, ∗∗∗p = 0.0007, ∗∗∗∗p < 0.0001, Kruskal-Wallis test with Dunn’s multiple comparison; n in C–H is the same as in A. All experiments are performed with wild type virgin females. In all scatterplots, each data point represents one fly, error bars indicate median with interquartile range.
Distribution of fru+ GABAergic neurons
Having established the functional relevance of fru+ GABAergic neurons for behavioral coordination and song patterning during male courtship, we next were interested to identify specific neurons responsible for the observed defects. Since the neurotransmitter identities of fru+ neurons have not been comprehensively mapped, we first identified all fru+ GABAergic neurons marked by coexpression of fru-GAL4 and GAD1-Lex47 transgenes (Figure S3A) in the different brain48 and ventral nerve cord49 regions. In the brain, fru+ GABAergic neurons were present in seven distinct clusters, some of which have been previously named and described50,51,52,53,54,55 (Figures S3B and S3C). In the ventral nerve cord (VNC), the cell bodies of fru+ GAD1+ neurons were less clustered and more variable than in the brain but could be grouped as belonging to four different regions (Figures S3B and S3D). We counted the anatomically defined neuronal classes/groups, which comprised 1–35 cells each, and found 130 ± 19.8 (n = 5, s.d.) neurons in the brain and 89.6 ± 20.5 (n = 8, s.d.) neurons in the VNC, that is, 11 subpopulations of approx. 220 fru+ GAD1+ neurons in the entire central nervous system. Functions of some fru+ GABAergic brain neurons are known, but neurons in the VNC have not been explored before (Table S1). For the anatomical assessment of fru+ Rdl+ neurons we visualized neurons coexpressing fru-FLP32 and Rdl-GAL447 transgenes (Figure S4). A large proportion of fru+ neurons express Rdl. Among them are neurons innervating prominent regions of the fru circuit32 such as the lateral protocerebral complex in the brain and the mesothoracic triangle in the VNC (Figure S4).
fru+ GABAergic neurons in the ventral nerve cord are required for song patterning
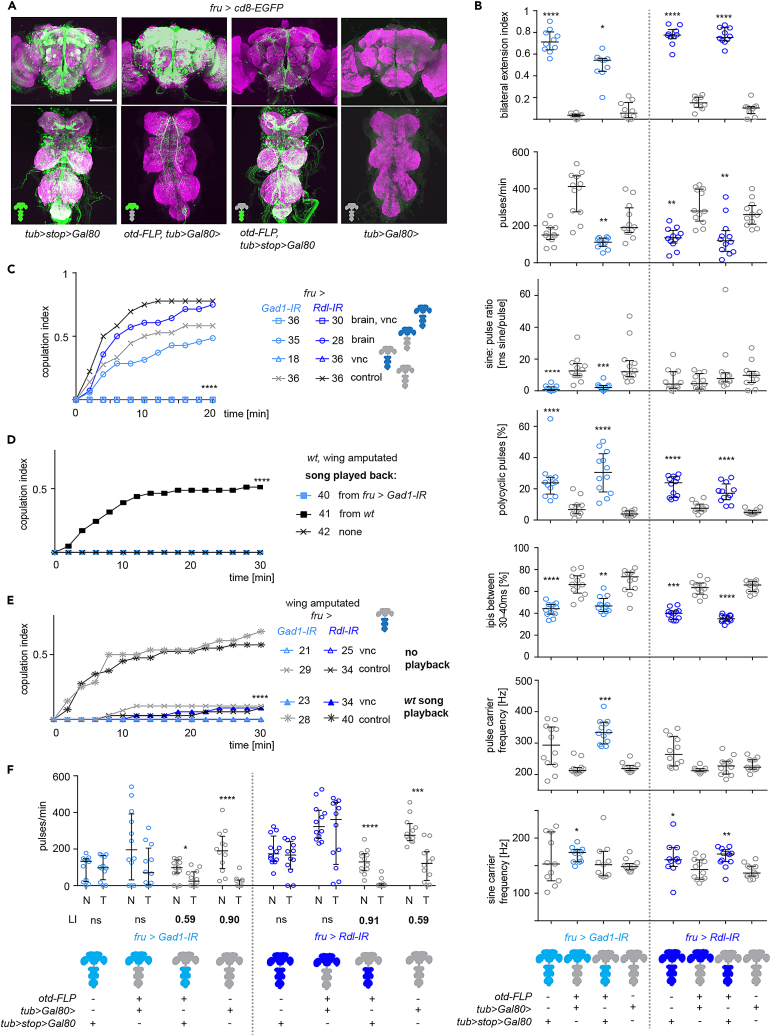
To distinguish between the functional roles of fru+ GABAergic neurons in the brain versus in the VNC, we restricted the knockdown of GAD1 and Rdl to each of the two tissues using the brain-specific otd-FLP transgene in combination with different FLP recognizable Gal80 cassettes. As visualized by membrane bound cd8-EGFP expression, this approach resulted in the respective expression domains, allowing us to probe fru+ neurons in the brain separately from fru+ neurons in the VNC (Figure 3A). When either GAD1 or Rdl was knocked down in the VNC only, flies displayed bilateral wing extension during singing. In contrast, brain-restricted knockdown did not lead to any bilaterality phenotype. Likewise, the alterations in acoustic song properties seen upon knockdown of GAD1 or Rdl in fru+ neurons of the entire nervous system were mostly replicated by VNC restricted, but not brain-restricted knockdown (Figure 3B). GAD1 and Rdl were required in fru+ VNC, but not brain neurons for short-term copulation success (Figure 3C). Given the changes in song structure upon the manipulation of fru+ VNC neurons, we wondered if the rejection of altered song by females was causing the observed defects in copulation success. To test this hypothesis, we experimentally separated song phenotype and male genotype by amputating both wings of wild type male flies and playing back song from GAD1 knockdown males or wild type male song while the amputated males courted virgin females. Wild type song, but not structurally changed song from fru > GAD1-IR knockdown males could rescue the short-term copulation success of wing-amputated males. Song from knockdown males was equally inefficient as no playback (silence), with both conditions leading to no copulations among 40 couples after 30 min (Figure 3D). This indicates that female receptivity, male courtship motivation, or a combination of both is not sufficiently stimulated by courtship song altered by GAD1 depletion. When wild type song was played back during courtship of wing-amputated males with GAD1 or Rdl knockdown in fru+ VNC neurons, these males remained unsuccessful in obtaining copulations (Figure 3E). We conclude that defect song upon GAD1 or Rdl knockdown in fru+ VNC neurons is sufficient to cause a defect in copulation success. However, fru+ GABAergic neurons in the VNC are also required for copulation in some other way, playing an additional, song-independent role. Males with GAD1 knockdown in either brain or VNC fru+ neurons only displayed a normal frequency of copulation attempts, in contrast to the increased copulation attempt frequency seen upon knockdown of GAD1 in the whole nervous system (Figure S5). VNC restricted, but not brain-restricted knockdown of GAD1 led to a decrease of the fraction of appropriate copulation attempts (Figure S5). For Rdl, an increased copulation attempt frequency seen upon knockdown in VNC neurons only, but not for knockdown in brain neurons. Knockdown of Rdl in either brain or VNC neurons only was sufficient to produce the phenotype of inappropriate copulation attempts (Figure S5). We wondered if phenotypes with decreased short-term copulation success showed a decreased frequency of copulation attempts when copulation attempts that we rated as inappropriate (i.e., with clearly visible, macroscopic mispositioning) were excluded from analysis. The copulation attempt indices excluding inappropriate attempts were not significantly decreased for any knockdown genotype as compared to controls and showed an increase upon knockdown of GAD1 or Rdl in fru+ neurons of the entire nervous system (Figure S5). In summary, the frequency of copulation attempts and the occurrence of clearly inappropriate attempts does not correlate directly with short term copulation success in the genotypes we tested here.
Figure 3.
Distinct roles of fru+ GABAergic neurons in brain and VNC
(A) Representative brain or VNC-restricted transgene expression in fru+ GABAergic neurons by combining brain-specific otd-FLP with different constructs of the GAL4 repressor GAL80 and genetic controls. Neuronal arbours in the VNC of the brain-specific otd-FLP, tub>stop>Gal80 specimen are from descending neurons, neuronal arbours in the brain of the VNC-specific otd-FLP, tub>Gal80> specimen are from ascending neurons and neurons in the gnathal ganglia. Cd8-EGFP expression in green, neuropil anti-bruchpilot staining in magenta, scale bar: 100 μm.
(B) Effects on song wing usage and song patterning of brain or VNC-specific RNAi-mediated knockdown of GAD1 or Rdl in fruitless neurons and respective genetic controls. n = 10 per genotype for bilateral extension index, n = 12 flies per genotype for song parameters. Full and partial knockdown genotypes are compared to the fully repressed tub>Gal80> controls, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001, Kruskal-Wallis test with Dunn’s multiple comparison.
(C) Copulation index of male flies upon RNAi-mediated knockdown of GAD1 or Rdl in fruitless neurons and respective genetic controls. ∗∗∗∗p < 0.0001, Fisher exact test.
(D) Copulation index of wing-amputated wt male flies supplemented with played back courtship song from a fru > GAD1-IR or wt male. ∗∗∗∗p < 0.0001, Fisher exact test.
(E) Copulation index of wing-amputated males with VNC-specific knockdown and controls with and without played back courtship song from wt male. ∗∗∗∗p < 0.0001, Fisher exact test.
(F) Effect of courtship conditioning in knockdown and control males. For each genotype the amount of song (in pulses/min) toward a mated wild type female is shown for naive (N) males and males trained (T) with an unreceptive mated wild type female. ∗p = 0.02, ∗∗∗∗p < 0.0001, Mann-Whitney test. The learning index of each genotype (LI) is given under the graph in the cases where significant learning occurred. n = 12–15 flies per group/genotype. (B–E) All experiments are performed with wild type virgin females. (C–E) n, number of flies tested, is indicated in the legend. (B–F) Blue colored parts in the nervous system cartoons indicate expression of the IR transgenes mediating knockdown. In B and F, genotypes with significant difference to the respective tub>Gal80> control (expression of IR transgene repressed in the whole nervous system) are indicated by blue colored data points. In all scatterplots, each data point represents one fly, error bars indicate median with interquartile range.
Lastly, suppression of singing after courtship conditioning depended on GAD1 and Rdl expression in fru+ brain neurons and was not affected by GAD1 or Rdl depletion in fru+ VNC neurons (Figure 3F).
In summary, different courtship and mating defects upon the depletion of GABAergic signaling in fru+ neurons, are due to different groups of neurons. Main song patterning defects (bilateral wing usage, reduction of sine song, polycyclicity, and broadening of the ipi range) and short-term copulation failure result from impairing GABAergic fru+ neurons in the VNC. Defects in courtship conditioning, i.e., loss of the reduction of song after training, result from impairing GABAergic fru+ neurons in the brain.
Discussion
Drosophila male courtship is an excellent model system of how complex behavior is orchestrated by a male-specific set of interconnected neurons, the fru+ circuit.32 Recently published connectomes of the Drosophila nervous system from electron microscopy (EM) reconstructions,56,57,58,59,60,61,62,63 will soon allow for in more detail the synaptic connectivity of identified fru+ neurons. Computational tools allow for the prediction of the neurotransmitter type from EM images,64 the analysis of large light microscopy expression datasets65,66 and the cross-comparison of EM and light microscopy data.67 In the face of this wealth of anatomical data, it is critical to mechanistically link the architecture of circuit motifs to behavior. Based on our analysis of the depletion of GABAergic signaling the fru+ circuits in brain and ventral nerve cord, we propose several hypotheses of how inhibitory motifs shape critical aspects of male courtship behavior and thus are required for mating success.
Inhibitory signaling is required for male copulation success
We find that RNAi-mediated knockdown of GAD1 or Rdl in fru+ neurons of the ventral nerve cord prevents the copulation of the manipulated male with a wild type female within 20 min of courtship. The absence of copulation within normal time spans could stem from either the lack of female receptivity or the lack of male courtship intensity, motor skill and coordination. We find indication that both is the case. The lack of inhibition alters the species-specific pattern of the courtship song to such an extent that playback of the acoustic signal, in contrast to control song, cannot rescue copulation defect of mute (wing amputated) wild type males (Figure 4D). If defect song was the main cause of preventing knockdown males from copulating, playback of wt control song to wing amputated animals should rescue the phenotype. We only observe such rescue in control males with functional GABAergic signaling (Figure 4D). Judged from the frequency of wing extensions and the number of copulation attempts (Figures 1B and 1E), knockdown of GAD1 or Rdl does not decrease overall courtship intensity, but slightly increases it. The males are clearly capable of bending their abdomen in the typical copulation attempt posture. However, they often attempt copulation when it is futile, i.e., when the female is far away or at an inappropriate angle (Figures 1C and 1D). During normal courtship, copulation attempts are tightly gated by the spatial relationship of the two sexes.20 We hypothesize that the disinhibition of copulation attempt command neurons, e.g., aSP2229 could allow the male to perform precisely timed attempts. A GABAergic interneuron receiving proximity information might inhibit a GABAergic gating neuron that is connected to the ventral nerve cord arbors of aSP22 (Figure 4A). Brain or VNC-specific knockdown experiment indicate that GABAergic signaling in both brain and VNC contribute to suppressing inappropriate copulation attempts (Figure S5). The proximity and positional information might be derived from both visual and olfactory stimuli. These modalities have previously been shown to act redundantly to guide the defined position of the courting male behind the female.68 When all clearly inappropriate copulation attempts are excluded from analysis, the frequency of remaining copulation attempts in brain or VNC knockdown males is the same as in control flies, or even higher when knockdown is performed in the whole nervous system (Figure S5). This indicates that defects more subtle than gross mispositioning prevent copulation success upon knockdown of GABAergic signaling in the VNC. In our experiments, we did not track at high temporal and spatial resolution the exact kinematics of copulation attempts as well as the movement of female genital plates and are cautious to define all not clearly inappropriate attempts as “appropriate” copulation attempts. It is therefore unclear if the higher frequency of copulation attempts in some knockdown genotypes (Figures 1B and S5) can be explained by the fact that males attempt a range of more or less subtly inappropriate attempts additionally to appropriate attempts. It seems likely that several modalities, computations, and inhibitory circuit motifs (encoding female speed, distance, heading, genital configuration) are combined to inhibit or disinhibit a copulation attempt. Broad removal of inhibition might lead to a broad mixture of attempts that are inhibited for different reasons by different neurons in control flies. Additionally, the general drive to attempt copulation in a suitable situation might be increased.
Figure 4.
Proposed inhibitory circuit motifs for aspects of courtship behavior
(A) Fast and precisely timed copulation attempts could arise by the disinhibition of the command neuron for copulation attempts, aSP22. Loss of inhibition leads to inappropriate copulation attempts.
(B) Circuit model for wing choice during song and its dependence on a circuit motif with reciprocal inhibition. Lateralized information about female proximity inhibits the ipsilateral b3 mn and disinhibits the contralateral b3 mn. Contralateral b3 mn and b3 muscle activation draws in the contralateral wing. The lateral inhibition motif allows for stable wing choice in the absence of strongly lateralized sensory input. Loss of inhibition leads to bilateral wing usage during singing.
(C) Model proposed by Roemschied et al.69 for the alternation of pulse and sine song, adapted to a version based on GAD1+ Rdl+ fru+ neurons. Both song modes are elicited by the pulse command neuron pIP10. Sine song neurons are active by rebound excitation after feedback inhibition has terminated a pulse song train. Loss of inhibition leads to loss of sine song.
(D) Restriction of pulse length by mechanosensory feedback from pulse start, activating a disinhibition motif gating i2 mn activity. i2 mn and i2 muscle activity stop wing movement.70,71 Loss of inhibition leads to long, polycyclic pulses.
(E) Restriction of ipi variability by the normalization of vPR6 activity. vPR6 activity is inversely correlated with ipi.28 Stabilization of vPR6 activity could arise from simple feedback or feedforward inhibition. Loss of inhibition leads to an a broadening of the ipi distribution. Expression of GAD1 is indicated in Cyan, expression of Rdl in Blue. aSP22, pIP10, vPR6 and the yet unidentified GAD1+ and GAD1+ Rdl+ cells depicted are fru+.
Under favorable conditions such as the presence of multiple virgin females on food, males with depleted GABAergic signaling in fru+ neurons manage to copulate after prolonged periods of courtship and can produce offspring (Figures S1A and S1B). Knockdown of GAD1 and Rdl has a different effect on copulation behavior and the timing of copulation. Knockdown of GAD1 leads to the occurrence of copulations lasting shorter than 8 min, in addition to normally timed copulations (Figure S1C). In contrast, flies depleted of Rdl in fru+ neurons do not show any short copulations and copulate longer than controls (Figure S1D). Since their fertility is decreased, a fraction of these long copulations might not be fertile. Previously, it has been shown that the silencing of GABAergic neurons expressing the sex determination factor Doublesex (Dsx) leads to the occurrence of prolonged copulations or copulations in which males were “stuck” and failed to uncouple from the female.72 A subset of these dsx+ GABAergic neurons that are sufficient for the prolonged copulation phenotype were shown not to express fru.73 dsx+ GABAergic neurons necessary for ending copulation in time might be antagonistic to the fru+ GABAergic neurons necessary to prevent premature uncoupling seen in short copulations. It is possible that the opposing phenotypes seen upon GAD1 and Rdl knockdown in fru+ neurons stem from the same class of neurons that shorten copulation when they lose their inhibitory activity (knockdown of GAD1) and prolong copulation when they lose inhibitory input (knockdown of Rdl).
GABAergic inhibition in the brain mediates courtship conditioning
GABAergic inhibition in fru+ circuit motifs in the brain has previously been suggested to act as gain control, normalizing the response to excitatory courtship stimuli.43,44 One might therefore predict excessive courtship upon the loss of inhibition. Interestingly, we do not see a large increase of courtship singing upon the depletion of GABAergic signaling (Figures 1E and 2B). Loss of inhibition however lead to a strong increase in the amount of song in a situation where control males sing little, i.e., after aversive conditioning with an unreceptive female. Courtship conditioning is often used as a paradigm for studying learning and memory.74,75 The inability of knockdown males to undergo courtship conditioning in our experiments can be interpreted as various, not mutually exclusive defects: the inability to perceive unsuccessful courtship as aversive, the inability to form a memory of the aversive experience and the inability to recruit the appropriate motor program (courtship suppression) upon retrieval of the memory. Since little is known about GABAergic signaling in the circuits for courtship conditioning, we here remain agnostic about the underlying causes of this phenotype.
GABAergic control of unilateral wing usage
Normal male courtship song is accompanied by unilateral wing extension. Depletion of GAD1 or Rdl leads to the prevalence of bilateral wing extension. Wing choice during courtship singing depends on both gustatory and visual input.27,52 During unilateral singing, the wing control muscle b3 is active on the side of the folded wing, suggesting that its innervating motor neuron b3 mn prevents the extension and full vibration of the wing not used in singing.18,71 We hypothesize that the reciprocal inhibition of Rdl+ GAD1+ fru+ interneurons that also inhibit b3 mn ensure unilateral wing usage (Figure 4B). These neurons are predicted to receive input from the above-mentioned sensory pathways.
A similar network motif with reciprocal inhibition has been shown to direct the lateralized escape response of zebrafish larva70 and might be a widespread architecture allowing behavioral choice during asymmetric motor behavior of bilateral animals.
Previously, the activity of brain dsx+ neurons has been suggested to restrict wing usage during singing from the bilateral to the unilateral pattern.76 In our experiments, we see that the depletion of VNC, but not brain GABAergic signaling in fru+ neurons gives rise to bilateral singing. Inactivation of dsx+ GABAergic neurons has been shown to affect copulation, but not earlier courtship steps such as singing behavior.72 We confirmed this finding and could not detect bilateral singing upon the silencing of dsx+ GABAergic neurons (data not shown). From this, we conclude that the fru+ GABAergic in the VNC that are necessary for unilateral wing usage are dsx-.
Courtship song patterning by GABAergic inhibition
Rhythmically structured motor programs support many essential behaviors such as locomotion, breathing and eating.77,78,79,80 They are also important in animal communication, conferring relevant information such as species identity and sender fitness to signals exchanged between conspecifics.81 Drosophila male song has an organized, precisely timed structure and serves as good model for studying circuit mechanisms underlying pattern generation.18 A small set of wing motor neurons has been shown to control main song parameters: the prevalence of the two song modes (sine song and pulses song), the length of pulses, and the spacing of the pulses (ipi).71 We find that inhibitory interneurons in the ventral nerve cord are required for sine song production, restriction of the pulse length to 1–2 wing beat cycles and the timing of the ipi (Figures 2A–2F and 3B). A few fru+ interneurons in the ventral nerve cord that affect song structure have been identified,28,82 but to our knowledge none of these identified neurons has been shown to be GABAergic. A recent model by Roemschied et al.69 suggests that inhibitory neurons are at the core of pulse and sine songs sequencing. A pair of neurons that reciprocally inhibit each other as well as either a pulse or a sine activating neuronal element ––could generate and alternation of both song modes (Figure 4C). Shiozaki et al.83 find that overlapping neuronal populations are active during sine and pulse song, with less neurons active during sine and additional neurons recruited during pulse. This could be interpreted as sine song relying on the inhibition of a pulse-specific interneuron population. In both models, loss of neuronal inhibition would lead to loss of sine, but not pulse song, as long as excitatory input from song promoting brain neurons is intact.
Additional to the severe reduction of sine song, we observe the appearance of pulse song with long, polycyclic pulses upon knockdown of GAD1 or Rdl (Figures 2A, 2D, and 3B). This phenotype resembles the one seen upon the silencing of the motor neuron innervating the wing control muscle i2, i2 mn. The i2 muscles has been proposed to act as a “pulse stopper,” actively terminating the vibration of the wing after 1–2 cycles of up- and downward movement.71 Moreover, it has been suggested that such a pulse stop mechanisms could arise from mechanosensory feedback.84 Here, we propose that the disinhibition of i2 mn by GAD1+ Rdl+ fru+ interneurons, activated by mechanosensory feedback from the initiation of the wing beat, prevents normal song pulses from escalating into a polycyclic pattern (Figure 4D). Such an escalation is observed when the inhibitory motif is impaired, since wing power muscles are stretch-activated and support longer wing oscillations once the thorax is deformed by a wing deflection.85
Wild type courtship pulses occur in trains where they are spaced at approximately 35 ms at 25°C. While in control song, around 80% of all inter pulse interval (ipis) are 30–40 ms long, depletion of GAD1 or Rdl leads to an ipi distribution that is more variable, but has a similar mode value (Figures 2E and 2F). Very little is known about circuit mechanism that control ipi. Possibly, ipi timing could be disrupted if it relies on correctly timed firing of neurons involved in the length and cyclicity of a single pulse. Previous work has shown that increasing activation strength of the fru+ interneuron vPR6 leads to decreased ipis of the elicited pulse trains.28 vPR6 does not express GAD1 (Figures S3A and S3B). If vPR6 firing rates must be within a certain range to maintain the normal ipi distribution, its output and/or excitatory input could be normalized and stabilized by simple feedback inhibition or feedforward inhibition circuit motifs (Figure 4E). Future identification of genetic lines for the specific manipulation of individual fru+ GABAergic neuronal classes among the around 90 ventral nerve cord cells (Table S1) will open the possibility to study song patterning in more detail.
Dsx expressing neurons in the VNC have been previously implicated in song patterning,82,86,87 leading to the question if any of our song phenotypes depends on fru+ GABAergic neurons that are also dsx+. No song phenotypes have been reported upon the silencing of all dsx+ GABAergic neurons.72 We confirmed this by assessing the song of the genotype described in Pavlou et al.72 (GAD1.AD, Dsx.DBD split GAL4 driving the expression of Tetanus toxin to silence neuronal activity, data not shown) and conclude that the fru+ GABAergic neurons shaping song structure are dsx-.
Nervous systems generate behavioral output in accordance with the internal state of the animal and process external stimuli relevant to this state. Neuronal inhibition enables them to do so in an efficient and adaptive way. GABAergic inhibition fine-tunes and organizes male Drosophila courtship behavior on many levels. Upon the depletion of inhibitory, males still perform all steps of the behavioral sequence in response to females. However, the motor execution is inaccurately timed and uncoordinated, severely reducing reproductive success. Further studies, helped by male nervous system connectomes, should identify the cellular identities of the fru+, GAD+, and/or Rdl+ interneurons underlying the here described behavioral phenotypes and test our proposed motif models for the gating of copulation attempts, unilateral wing, sequencing of sine and pulse song, and pulse timing.
Limitations of the study
The phenotypes described in this study were not linked to single identifiable cell types. Moreover, we cannot ascertain the level of RNAi-mediated knockdown of GAD1 and Rdl expression in the different fru+ neurons. Phenotypes might arise from a reduction or a complete loss of GAD1/Rdl protein.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GFP antibody (rabbit) | Torrey Pines Biolabs | Cat# TP401 071519; RRID:AB_10013661 |
| anti-Gfp antibody (chicken) | Abcam | RRID: AB_300789 |
| anti-DsRed antibody (rabbit) | Living colors/Takara Bio | RRID: AB_10013483 |
| nc82/ bruchpilot antibody (mouse) | Developmental Studies Hybridoma Bank |
Cat# nc82; RRID:AB_2314866 |
| Goat anti-Mouse IgG (H+L) Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21236; RRID:AB_2535805 |
| Goat anti-Rabbit IgG (H+L) Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11034; RRID:AB_2576217 |
| Goat anti-Rabbit IgG (H+L) Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A-11011; RRID:AB_143157 |
| Chemicals, peptides, and recombinant proteins | ||
| Vectashield | Vector Labs | Cat# H-1000 |
| Experimental models: Organisms/strains | ||
| Drosophila: fruitless-GAL4 (fru-GAL4) | Barry Dickson | BDSC_66696 |
| Drosophila: y v; P{TRiP.HMC03350}attP40 (UAS-IR Gad1,Gad1-IR) | Bloomington Drosophila Stock Center (BDSC) |
BDSC_51794 |
| Drosophila: w1118; P{GD8508}v32344 (UAS-IR Gad1∗) | Vienna Drosophila Resource Center (VDRC) | SCR_013805 |
| Drosophila: y sc v sev; P{TRiP.HMC03643}attP40 (UAS-IR Rdl, Rdl-IR) | BDSC | BDSC_52903 |
| Drosophila: w1118; P{GD4609}v41103 (UAS-IR Rdl∗) | VDRC | SCR_013805 |
| Drosophila: P{KK101500}VIE-260B (UAS-IR vGAT) | VDRC | SCR_013805 |
| Drosophila: Wild type Canton S (wt) | Barry Dickson | n.a. |
| Drosophila: w1118, P{UAS-dicer2, w[+]} (UAS-Dcr2) | VDRC | SCR_013805 |
| Drosophila: w-; UAS-mCD8-Egfp; + (cd8-EGFP) | Barry Dickson | n.a. |
| Drosophila: w-; LexAop-mCD8-tomato; + (cd8-Tomato) | Barry Dickson | n.a. |
| Drosophila: w; Mi{Trojan-lexA:QFAD.2}Gad1[MI09277-TlexA:QFAD.2] (Gad1-LexA) | Ben White | BDSC_60324 |
| Drosophila: UAS-FRT-stop-FRT-mCD8-Egfp (UAS>stop>cd8-EGFP) | Barry Dickson | n.a. |
| Drosophila: w-; ;TI{FLP}fru[FLP]/TM3, Sb (fru-FLP) | Barry Dickson | BDSC_66870 |
| Drosophila: Mi{Trojan-GAL4.1}Rdl[MI02957-TG4.1] (Rdl-GAL4) | Ben White | BDSC_65421 |
| Drosophila: w; Otd-nlsFLPo.attp40; + (Otd-FLP) | David Anderson | n.a. |
| Drosophila: w; +; tubP-FRT-stop-FRT-GAL80 (tub>stop>Gal80) | BDSC | BDSC_39213 |
| Drosophila: w; +; tubP-FRT-GAL80-FRT (tub>Gal80>) | Barry Dickson | n.a. |
| Software and algorithms | ||
| Zen | Carl Zeiss Microscopy | RRID:SCR_013672 |
| FlySongSegmenter | Benjamin Arthur and David Stern | https://github.com/FlyCourtship/FlySongSegmenter |
| MATLAB | MathWorks | RRID:SCR_001622 |
| Graph Pad Prism6 | GraphPad Software | RRID:SCR_002798 |
| Fiji | https://fiji.sc/ | RRID: SCR_002285 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Anne C. von Philipsborn (anne.vonphilipsborn@unifr.ch).
Materials availability
This study did not create new unique reagents.
Experimental model and study participant details
Drosophila melanogaster transgenic lines and stock maintenance
Drosophila lines used and their origin are listed in the key resources table. For control genotypes, GAL4 and UAS lines were crossed with wild type flies. For RNAi mediated knockdown with lines from the VDRC libraries, UAS-Dcr2 was used to enhance knockdown efficiency. Flies were grown on regular food (water, cornmeal, oatmeal, sucrose, yeast, agar, acetic acid, preservative methyl-4-hydroxybenzoate) at 25°C, 60% humidity, and a 12h/12h light-dark cycle.
Method details
Behavior assays
For all behavioral assays, if not otherwise indicated, males were individually aged for 4–7 days and paired with 4–7 d old CS virgins that were aged in groups of 10–20 flies. All courtship assays were performed within 3 hrs after lights on or 3 hrs before lights off (activity peaks) at 25°C, 60% humidity. The genotypes of the experimental flies (all males) are listed in Table S2.
Each experiment was replicated 2–3 times by repeating the respective cross and recording the behavior of each genotype on several different days. Video and audio files were evaluated with the observer blind to the genotype of the animals.
Single pair courtship assays
Single pair courtship assays were performed as described previously.88 For evaluating copulation success, pairs were videotaped for 20 min in chambers of 1 cm diameter. For evaluating wing extension and copulation attempts, pairs were videotaped in chamber of 1.7 cm diameter with beveled walls, which prevented flies from walking at the side walls of the chambers and allowed for continuous top view. Wing extensions were scored for the first 5 min, every 2.5 s, i.e., in approximately 120 frames per pair. The wing extension index was calculated as the fraction of frames in which the male extended at least one wing at an angle of more than 30°. The bilateral extension index was calculated as the fraction of wing extensions during which the male extended both wings at an angle of more than 30°. The large angle extension index was calculated as the fraction of wing extensions during which the male extended at least one wing at an angle of more than 85°. Copulation attempts were scored for the first 2 min, every 0.5 s, i.e., in approximately 120 frames per pair. The copulation attempt index is the fraction of frames in which the male attempted copulation, characterized by pronounced abdominal bending. The appropriate copulation attempt index is the fraction of copulation attempts during which the male pointed his abdomen toward the female genitalia and was not further than 1 fly length away from the female.
Courtship suppression assays
For courtship suppression assays,89 males were paired for 1 hr with a mated CS female in a chamber of 1 cm diameter (trained group) or left alone for 1 hr in the chamber (naïve group). Males were then transferred into song recording chambers with a new mated female and courtship song was recorded for 4 min. Learning and suppression of song was judged to have occurred when the amount of pulse song was significantly different in trained and naïve groups.
Fertility and long-term mating assays
For the assessment of fertility, a single male fly ways paired with three wild type virgin flies in a food vial for three days. After 10 further days, the presence of offspring (larvae) was evaluated. For long term mating assays, a single male fly ways paired with three wild type virgin flies in a food vial with a sloped surface. Vials were imaged by time lapse video recording (1 frame/s) for 12hrs.
Courtship song recording
Song recording and evaluation was performed as described previously.71,90 In brief, we used a multi-channel array designed by the Stern lab, detected pulse and sine song with a custom MATLAB script91 and used visualization of oscillograms to manually correct automated detection. Each datapoint corresponds to an approximately 4 min long recording of one male fly.
Playback experiments
Playback experiments were performed as described previously.90,92 Wings were amputated with spring scissors close to the hinge and flies were allowed to recover for at least 24 hrs before testing. The approximately 3 s long song examples from a wt and fru > Gad1-IR fly were continuously played back and had the following characteristics representative for their genotype: number of pulses; 38 and 34; % ipis between 30-40 ms: 86 and 41; % polycyclic pulses 3 and 41; median pulse frequency: 246 Hz and 322 Hz, respectively.
Immunohistochemistry and confocal imaging
Immunohistochemistry and confocal imaging of fly nervous systems were performed as described previously.71 Fly nervous systems were dissected in PBS, fixed in 4% paraformaldehyde, stained with mouse nc82 antibody (Developmental Studies Hybridoma Bank, RRID: AB_2314866), rabbit anti-GFP antibody (Torrey Pines Biolabs, RRID: AB_10013661) or chicken anti-Gfp antibody (Abcam, RRID: AB_300789), rabbit anti-DsRed antibody (Living colors/Takara Bio, RRID: AB_10013483), followed by respective secondary antibodys (Goat IgG (H+L) Alexa Fluor 488, 568 and 647) and mounted in Vectashield (Vector labs, RRID: AB_2336789). Confocal image stacks were acquired at a Zeiss LSM780 microscope.
Quantification and statistical analysis
General statistical analysis
For statistical test, Graph Pad prism software was used if not otherwise indicated. The tests used, test outcomes, sample size and details of what each datapoint represents can be found in the figure legends.
Learning index
The learning index (LI) for each genotype was calculated as the relative reduction of pulse song in trained males as compared to naïve males: (mean (pulses/min)Naive- mean (pulses/min)Trained )/ mean (pulses/min)Naïve. To test for differences between the LIs of two genotypes, pulse/min values from individual flies from trained groups were permuted between genotypes, and the same was done for naïve groups (1000000 permutations). The p value gives the probability for obtaining a difference in LIs equal to or larger than the experimentally observed one.
Courtship song parameters
The sine: pulse ratio was calculated by dividing the total amount of sine song in ms by the total number of pulses. Pulse cyclicity was measured as the minimum of positive and negative pulse peaks, counting all peaks with at least 2/3 the amplitude of the maximum peak within a pulse. A pulse was considered to be polycyclic when it had three or more peaks. For analysis of ipis, we evaluated only ipis between 15-80 ms in trains of more than 2 pulses. Carrier frequencies of pulse and sine song were assessed by plotting the median value of each fly.
Acknowledgments
We thank Asia Ahmed, Akhil John, Francesca Barbieri, Peter Kerwin, and Yanan Zhang for help with experiments, Anna Prudnikova, Marketa Kaderakova, Tatiana Adamiec, and Laurence Clément for technical assistance. We thank DGRC and VDRC stock libraries, Developmental Studies Hybridoma Bank for the nc82 antibody, Barry Dickson, and Ben White for providing fly stocks. This study was supported by Lundbeckfonden grant DANDRITE-R248-2016-2518 and by Swiss National Science foundation grant 310030_212222.
Author contributions
Conceptualization, methodology, visualization, supervision, and project administration, H.A., S.S.N, and A.C.v.P.; investigation and data curation, H.A., S.S.N, B.S., and A.C.v.P.; writing and funding acquisition, A.C.v.P.
Declaration of interests
The authors declare no competing interest.
Published: September 28, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108069.
Supplemental information
Data and code availability
All raw data is available and will be shared upon request to the lead contact.
This paper did not generate original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Rentzsch F., Juliano C., Galliot B. Modern genomic tools reveal the structural and cellular diversity of cnidarian nervous systems. Curr. Opin. Neurobiol. 2019;56:87–96. doi: 10.1016/j.conb.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Arendt D. The Evolutionary Assembly of Neuronal Machinery. Curr. Biol. 2020;30:R603–R616. doi: 10.1016/j.cub.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Pierobon P. An Interesting Molecule: γ-Aminobutyric Acid. What Can We Learn from Hydra Polyps? Brain Sci. 2021;11:437. doi: 10.3390/brainsci11040437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cossart R., Bernard C., Ben-Ari Y. Multiple facets of GABAergic neurons and synapses: multiple fates of GABA signalling in epilepsies. Trends Neurosci. 2005;28:108–115. doi: 10.1016/j.tins.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Hennequin G., Agnes E.J., Vogels T.P. Inhibitory Plasticity: Balance, Control, and Codependence. Annu. Rev. Neurosci. 2017;40:557–579. doi: 10.1146/annurev-neuro-072116-031005. [DOI] [PubMed] [Google Scholar]
- 6.Yu L., Yu Y. Energy-efficient neural information processing in individual neurons and neuronal networks. J. Neurosci. Res. 2017;95:2253–2266. doi: 10.1002/jnr.24131. [DOI] [PubMed] [Google Scholar]
- 7.Smart T.G., Stephenson F.A. A half century of γ-aminobutyric acid. Brain Neurosci. Adv. 2019;3 doi: 10.1177/2398212819858249. 2398212819858249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadeh S., Clopath C. Inhibitory stabilization and cortical computation. Nat. Rev. Neurosci. 2021;22:21–37. doi: 10.1038/s41583-020-00390-z. [DOI] [PubMed] [Google Scholar]
- 9.Braganza O., Beck H. The Circuit Motif as a Conceptual Tool for Multilevel Neuroscience. Trends Neurosci. 2018;41:128–136. doi: 10.1016/j.tins.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Huang Z.J., Paul A. The diversity of GABAergic neurons and neural communication elements. Nat. Rev. Neurosci. 2019;20:563–572. doi: 10.1038/s41583-019-0195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baca S.M., Marin-Burgin A., Wagenaar D.A., Kristan W.B. Widespread Inhibition Proportional to Excitation Controls the Gain of a Leech Behavioral Circuit. Neuron. 2008;57:276–289. doi: 10.1016/j.neuron.2007.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudry Q., Kristan W.B. Behavioral choice by presynaptic inhibition of tactile sensory terminals. Nat. Neurosci. 2009;12:1450–1457. doi: 10.1038/nn.2400. [DOI] [PubMed] [Google Scholar]
- 13.Hangya B., Pi H.-J., Kvitsiani D., Ranade S.P., Kepecs A. From circuit motifs to computations: mapping the behavioral repertoire of cortical interneurons. Curr. Opin. Neurobiol. 2014;26:117–124. doi: 10.1016/j.conb.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swanson O.K., Maffei A. From Hiring to Firing: Activation of Inhibitory Neurons and Their Recruitment in Behavior. Front. Mol. Neurosci. 2019;12:168. doi: 10.3389/fnmol.2019.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ai H., Kumaraswamy A., Kohashi T., Ikeno H., Wachtler T. Inhibitory Pathways for Processing the Temporal Structure of Sensory Signals in the Insect Brain. Front. Psychol. 2018;9:1517. doi: 10.3389/fpsyg.2018.01517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jovanic T., Schneider-Mizell C.M., Shao M., Masson J.-B., Denisov G., Fetter R.D., Mensh B.D., Truman J.W., Cardona A., Zlatic M. Competitive Disinhibition Mediates Behavioral Choice and Sequences in Drosophila. Cell. 2016;167:858–870.e19. doi: 10.1016/j.cell.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Ellendersen B.E., von Philipsborn A.C. Neuronal modulation of D. melanogaster sexual behaviour. Curr. Opin. Insect Sci. 2017;24:21–28. doi: 10.1016/j.cois.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Swain B., von Philipsborn A.C. In: Advances in Insect Physiology. Jurenka R., editor. Academic Press; 2021. Chapter Three - Sound production in Drosophila melanogaster: Behaviour and neurobiology; pp. 141–187. [DOI] [Google Scholar]
- 19.von Philipsborn A.C., Shohat-Ophir G., Rezaval C. Measurement of Drosophila Reproductive Behaviors. Cold Spring Harb. Protoc. 2023;2023 doi: 10.1101/pdb.top107866. pdb.top107866. [DOI] [PubMed] [Google Scholar]
- 20.Mezzera C., Brotas M., Gaspar M., Pavlou H.J., Goodwin S.F., Vasconcelos M.L. Ovipositor Extrusion Promotes the Transition from Courtship to Copulation and Signals Female Acceptance in Drosophila melanogaster. Curr. Biol. 2020;30:3736–3748.e5. doi: 10.1016/j.cub.2020.06.071. [DOI] [PubMed] [Google Scholar]
- 21.Wang K., Wang F., Forknall N., Yang T., Patrick C., Parekh R., Dickson B.J. Neural circuit mechanisms of sexual receptivity in Drosophila females. Nature. 2021;589:577–581. doi: 10.1038/s41586-020-2972-7. [DOI] [PubMed] [Google Scholar]
- 22.Auer T.O., Benton R. Sexual circuitry in Drosophila. Curr. Opin. Neurobiol. 2016;38:18–26. doi: 10.1016/j.conb.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Rings A., Goodwin S.F. To court or not to court - a multimodal sensory decision in Drosophila males. Curr. Opin. Insect Sci. 2019;35:48–53. doi: 10.1016/j.cois.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Peng Q., Chen J., Pan Y. From fruitless to sex: On the generation and diversification of an innate behavior. Genes Brain Behav. 2021;20:e12772. doi: 10.1111/gbb.12772. [DOI] [PubMed] [Google Scholar]
- 25.Stockinger P., Kvitsiani D., Rotkopf S., Tirián L., Dickson B.J. Neural Circuitry that Governs Drosophila Male Courtship Behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 26.Manoli D.S., Foss M., Villella A., Taylor B.J., Hall J.C., Baker B.S. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436:395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- 27.Ribeiro I.M.A., Drews M., Bahl A., Machacek C., Borst A., Dickson B.J. Visual Projection Neurons Mediating Directed Courtship in Drosophila. Cell. 2018;174:607–621.e18. doi: 10.1016/j.cell.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 28.von Philipsborn A.C., Liu T., Yu J.Y., Masser C., Bidaye S.S., Dickson B.J. Neuronal Control of Drosophila Courtship Song. Neuron. 2011;69:509–522. doi: 10.1016/j.neuron.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 29.McKellar C.E., Lillvis J.L., Bath D.E., Fitzgerald J.E., Cannon J.G., Simpson J.H., Dickson B.J. Threshold-Based Ordering of Sequential Actions during Drosophila Courtship. Curr. Biol. 2019;29:426–434.e6. doi: 10.1016/j.cub.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Billeter J.C., Goodwin S.F. Characterization of Drosophila fruitless-gal4 transgenes reveals expression in male-specific fruitless neurons and innervation of male reproductive structures. J. Comp. Neurol. 2004;475:270–287. doi: 10.1002/cne.20177. [DOI] [PubMed] [Google Scholar]
- 31.Kimura K.I., Ote M., Tazawa T., Yamamoto D. Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature. 2005;438:229–233. doi: 10.1038/nature04229. [DOI] [PubMed] [Google Scholar]
- 32.Yu J.Y., Kanai M.I., Demir E., Jefferis G.S.X.E., Dickson B.J. Cellular Organization of the Neural Circuit that Drives Drosophila Courtship Behavior. Curr. Biol. 2010;20:1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 33.Cachero S., Ostrovsky A.D., Yu J.Y., Dickson B.J., Jefferis G.S.X.E. Sexual Dimorphism in the Fly Brain. Curr. Biol. 2010;20:1589–1601. doi: 10.1016/j.cub.2010.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Philipsborn A.C., Jörchel S., Tirian L., Demir E., Morita T., Stern D.L., Dickson B.J. Cellular and Behavioral Functions of fruitless Isoforms in Drosophila Courtship. Curr. Biol. 2014;24:242–251. doi: 10.1016/j.cub.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson F.R., Newby L.M., Kulkarni S.J. Drosophila GABAergic Systems: Sequence and Expression of Glutamic Acid Decarboxylase. J. Neurochem. 1990;54:1068–1078. doi: 10.1111/j.1471-4159.1990.tb02359.x. [DOI] [PubMed] [Google Scholar]
- 36.Ffrench-Constant R.H., Mortlock D.P., Shaffer C.D., MacIntyre R.J., Roush R.T. Molecular cloning and transformation of cyclodiene resistance in Drosophila: an invertebrate gamma-aminobutyric acid subtype A receptor locus. Proc. Natl. Acad. Sci. USA. 1991;88:7209–7213. doi: 10.1073/pnas.88.16.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen R., Belelli D., Lambert J.J., Peters J.A., Reyes A., Lan N.C. Cloning and functional expression of a Drosophila gamma-aminobutyric acid receptor. Proc. Natl. Acad. Sci. USA. 1994;91:6069–6073. doi: 10.1073/pnas.91.13.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison J.B., Chen H.H., Sattelle E., Barker P.J., Huskisson N.S., Rauh J.J., Bai D., Sattelle D.B. Immunocytochemical mapping of a C-terminus anti-peptide antibody to the GABA receptor subunit, RDL in the nervous system in Drosophila melanogaster. Cell Tissue Res. 1996;284:269–278. doi: 10.1007/s004410050587. [DOI] [PubMed] [Google Scholar]
- 39.Enell L., Hamasaka Y., Kolodziejczyk A., Nässel D.R. gamma-Aminobutyric acid (GABA) signaling components in Drosophila: Immunocytochemical localization of GABA(B) receptors in relation to the GABA(A) receptor subunit RDL and a vesicular GABA transporter. J. Comp. Neurol. 2007;505:18–31. doi: 10.1002/cne.21472. [DOI] [PubMed] [Google Scholar]
- 40.Croset V., Treiber C.D., Waddell S. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. Elife. 2018;7:e34550. doi: 10.7554/eLife.34550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davie K., Janssens J., Koldere D., De Waegeneer M., Pech U., Kreft Ł., Aibar S., Makhzami S., Christiaens V., Bravo González-Blas C., et al. A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell. 2018;174:982–998.e20. doi: 10.1016/j.cell.2018.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allen A.M., Neville M.C., Birtles S., Croset V., Treiber C.D., Waddell S., Goodwin S.F. A single-cell transcriptomic atlas of the adult Drosophila ventral nerve cord. Elife. 2020;9:e54074. doi: 10.7554/eLife.54074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kallman B.R., Kim H., Scott K. Excitation and inhibition onto central courtship neurons biases Drosophila mate choice. Elife. 2015;4:e11188. doi: 10.7554/eLife.11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clowney E.J., Iguchi S., Bussell J.J., Scheer E., Ruta V. Multimodal Chemosensory Circuits Controlling Male Courtship in Drosophila. Neuron. 2015;87:1036–1049. doi: 10.1016/j.neuron.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorter J.A., Jagadeesh S., Gahr C., Boonekamp J.J., Levine J.D., Billeter J.-C. The nutritional and hedonic value of food modulate sexual receptivity in Drosophila melanogaster females. Sci. Rep. 2016;6:19441. doi: 10.1038/srep19441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilchrist A.S., Partridge L. Why it is difficult to model sperm displacement in Drosophila melanogaster: The relation between sperm transfer and copulation duration. Evolution. 2000;54:534–542. doi: 10.1111/j.0014-3820.2000.tb00056.x. [DOI] [PubMed] [Google Scholar]
- 47.Diao F., Ironfield H., Luan H., Diao F., Shropshire W.C., Ewer J., Marr E., Potter C.J., Landgraf M., White B.H. Plug-and-Play Genetic Access to Drosophila Cell Types using Exchangeable Exon Cassettes. Cell Rep. 2015;10:1410–1421. doi: 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito K., Shinomiya K., Ito M., Armstrong J.D., Boyan G., Hartenstein V., Harzsch S., Heisenberg M., Homberg U., Jenett A., et al. A Systematic Nomenclature for the Insect Brain. Neuron. 2014;81:755–765. doi: 10.1016/j.neuron.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 49.Court R., Namiki S., Armstrong J.D., Börner J., Card G., Costa M., Dickinson M., Duch C., Korff W., Mann R., et al. A Systematic Nomenclature for the Drosophila Ventral Nerve Cord. Neuron. 2020;107:1071–1079.e2. doi: 10.1016/j.neuron.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruta V., Datta S.R., Vasconcelos M.L., Freeland J., Looger L.L., Axel R. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468:686–690. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- 51.Koganezawa M., Kimura K.-I., Yamamoto D. The Neural Circuitry that Functions as a Switch for Courtship versus Aggression in Drosophila Males. Curr. Biol. 2016;26:1395–1403. doi: 10.1016/j.cub.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 52.Koganezawa M., Haba D., Matsuo T., Yamamoto D. The Shaping of Male Courtship Posture by Lateralized Gustatory Inputs to Male-Specific Interneurons. Curr. Biol. 2010;20:1–8. doi: 10.1016/j.cub.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 53.Liang L., Li Y., Potter C.J., Yizhar O., Deisseroth K., Tsien R.W., Luo L. GABAergic Projection Neurons Route Selective Olfactory Inputs to Specific Higher-Order Neurons. Neuron. 2013;79:917–931. doi: 10.1016/j.neuron.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan Q., Song Y., Yang C.-H., Jan L.Y., Jan Y.N. Female contact modulates male aggression via a sexually dimorphic GABAergic circuit in Drosophila. Nat. Neurosci. 2014;17:81–88. doi: 10.1038/nn.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaughan A.G., Zhou C., Manoli D.S., Baker B.S. Neural Pathways for the Detection and Discrimination of Conspecific Song in D. melanogaster. Curr. Biol. 2014;24:1039–1049. doi: 10.1016/j.cub.2014.03.048. [DOI] [PubMed] [Google Scholar]
- 56.Zheng Z., Lauritzen J.S., Perlman E., Robinson C.G., Nichols M., Milkie D., Torrens O., Price J., Fisher C.B., Sharifi N., et al. A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell. 2018;174:730–743.e22. doi: 10.1016/j.cell.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phelps J.S., Hildebrand D.G.C., Graham B.J., Kuan A.T., Thomas L.A., Nguyen T.M., Buhmann J., Azevedo A.W., Sustar A., Agrawal S., et al. Reconstruction of motor control circuits in adult Drosophila using automated transmission electron microscopy. Cell. 2021;184:759–774.e18. doi: 10.1016/j.cell.2020.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheffer L.K., Xu C.S., Januszewski M., Lu Z., Takemura S.-Y., Hayworth K.J., Huang G.B., Shinomiya K., Maitlin-Shepard J., Berg S., et al. A connectome and analysis of the adult Drosophila central brain. Elife. 2020;9:e57443. doi: 10.7554/eLife.57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Azevedo A., Lesser E., Mark B., Phelps J., Elabbady L., Kuroda S., Sustar A., Moussa A., Kandelwal A., Dallmann C.J., et al. Tools for comprehensive reconstruction and analysis of Drosophila motor circuits. bioRxiv. 2022 doi: 10.1101/2022.12.15.520299. Preprint at. [DOI] [Google Scholar]
- 60.Lesser E., Azevedo A.W., Phelps J.S., Elabbady L., Cook A., Mark B., Kuroda S., Sustar A., Moussa A., Dallmann C.J., et al. Synaptic architecture of leg and wing motor control networks in Drosophila. bioRxiv. 2023 doi: 10.1101/2023.05.30.542725. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takemura S.-Y., Hayworth K.J., Huang G.B., Januszewski M., Lu Z., Marin E.C., Preibisch S., Xu C.S., Bogovic J., Champion A.S., et al. A Connectome of the Male Drosophila Ventral Nerve Cord. bioRxiv. 2023 doi: 10.1101/2023.06.05.543757. Preprint at. [DOI] [Google Scholar]
- 62.Marin E.C., Morris B.J., Stuerner T., Champion A.S., Krzeminski D., Badalamente G., Gkantia M., Dunne C.R., Eichler K., Takemura S.-Y., et al. Systematic annotation of a complete adult male Drosophila nerve cord connectome reveals principles of functional organisation. bioRxiv. 2023 doi: 10.1101/2023.06.05.543407. Preprint at. [DOI] [Google Scholar]
- 63.Cheong H.S.J., Eichler K., Stuerner T., Asinof S.K., Champion A.S., Marin E.C., Oram T.B., Sumathipala M., Venkatasubramanian L., Namiki S., et al. Transforming descending input into behavior: The organization of premotor circuits in the Drosophila Male Adult Nerve Cord connectome. bioRxiv. 2023 doi: 10.1101/2023.06.07.543976. Preprint at. [DOI] [Google Scholar]
- 64.Eckstein N., Bates A.S., Du M., Hartenstein V., Jefferis G.S.X.E., Funke J. Neurotransmitter Classification from Electron Microscopy Images at Synaptic Sites in Drosophila. bioRxiv. 2020 doi: 10.1101/2020.06.12.148775. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Panser K., Tirian L., Schulze F., Villalba S., Jefferis G.S.X.E., Bühler K., Straw A.D. Automatic Segmentation of Drosophila Neural Compartments Using GAL4 Expression Data Reveals Novel Visual Pathways. Curr. Biol. 2016;26:1943–1954. doi: 10.1016/j.cub.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ehrhardt E., Whitehead S.C., Namiki S., Minegishi R., Siwanowicz I., Feng K., Otsuna H., FlyLight Project Team. Meissner G.W., Stern D., et al. Single-cell type analysis of wing premotor circuits in the ventral nerve cord of Drosophila melanogaster. bioRxiv. 2023 doi: 10.1101/2023.05.31.542897. Preprint at. [DOI] [Google Scholar]
- 67.Costa M., Manton J.D., Ostrovsky A.D., Prohaska S., Jefferis G.S.X.E. NBLAST: Rapid, Sensitive Comparison of Neuronal Structure and Construction of Neuron Family Databases. Neuron. 2016;91:293–311. doi: 10.1016/j.neuron.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kimura K.i., Sato C., Yamamoto K., Yamamoto D. From the back or front: The courtship position is a matter of smell and sight in Drosophila melanogaster males. J. Neurogenet. 2015;29:18–22. doi: 10.3109/01677063.2014.968278. [DOI] [PubMed] [Google Scholar]
- 69.Roemschied F.A., Pacheco D.A., Ireland E.C., Li X., Aragon M.J., Pang R., Murthy M. Flexible Circuit Mechanisms for Context-Dependent Song Sequencing. bioRxiv. 2021 doi: 10.1101/2021.11.01.466727. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koyama M., Minale F., Shum J., Nishimura N., Schaffer C.B., Fetcho J.R. A circuit motif in the zebrafish hindbrain for a two alternative behavioral choice to turn left or right. Elife. 2016;5:e16808. doi: 10.7554/eLife.16808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Sullivan A., Lindsay T., Prudnikova A., Erdi B., Dickinson M., von Philipsborn A.C. Multifunctional Wing Motor Control of Song and Flight. Curr. Biol. 2018;28:2705–2717.e4. doi: 10.1016/j.cub.2018.06.038. [DOI] [PubMed] [Google Scholar]
- 72.Pavlou H.J., Lin A.C., Neville M.C., Nojima T., Diao F., Chen B.E., White B.H., Goodwin S.F. Neural circuitry coordinating male copulation. Elife. 2016;5:e20713. doi: 10.7554/eLife.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crickmore M.A., Vosshall L.B. Opposing Dopaminergic and GABAergic Neurons Control the Duration and Persistence of Copulation in Drosophila. Cell. 2013;155:881–893. doi: 10.1016/j.cell.2013.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Griffith L.C., Ejima A. Courtship learning in Drosophila melanogaster: Diverse plasticity of a reproductive behavior. Learn. Mem. 2009;16:743–750. doi: 10.1101/lm.956309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raun N., Jones S., Kramer J.M. Conditioned courtship suppression in Drosophila melanogaster. J. Neurogenet. 2021;35:154–167. doi: 10.1080/01677063.2021.1873323. [DOI] [PubMed] [Google Scholar]
- 76.Pan Y., Robinett C.C., Baker B.S. Turning Males On: Activation of Male Courtship Behavior in Drosophila melanogaster. PLoS One. 2011;6:e21144. doi: 10.1371/journal.pone.0021144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kiehn O., Kullander K. Central pattern generators deciphered by molecular genetics. Neuron. 2004;41:317–321. doi: 10.1016/s0896-6273(04)00042-x. [DOI] [PubMed] [Google Scholar]
- 78.Sakurai A., Katz P.S. Phylogenetic and individual variation in gastropod central pattern generators. J. Comp. Physiol. 2015;201:829–839. doi: 10.1007/s00359-015-1007-6. [DOI] [PubMed] [Google Scholar]
- 79.Paton J.J., Buonomano D.V. The Neural Basis of Timing: Distributed Mechanisms for Diverse Functions. Neuron. 2018;98:687–705. doi: 10.1016/j.neuron.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mantziaris C., Bockemühl T., Büschges A. Central pattern generating networks in insect locomotion. Dev. Neurobiol. 2020;80:16–30. doi: 10.1002/dneu.22738. [DOI] [PubMed] [Google Scholar]
- 81.Greenfield M.D. Oxford University Press; 2002. Signalers and Receivers: Mechanisms and Evolution of Arthropod Communication. [Google Scholar]
- 82.Shirangi T.R., Wong A.M., Truman J.W., Stern D.L. Doublesex Regulates the Connectivity of a Neural Circuit Controlling Drosophila Male Courtship Song. Dev. Cell. 2016;37:533–544. doi: 10.1016/j.devcel.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 83.Shiozaki H.M., Wang K., Lillvis J.L., Xu M., Dickson B.J., Stern D.L. Neural coding of distinct motor patterns during Drosophila courtship song. bioRxiv. 2022 doi: 10.1101/2022.12.14.520499. Preprint at. [DOI] [Google Scholar]
- 84.Ewing A.W. The role of feedback during singing and flight in Drosophila melanogaster. Physiol. Entomol. 1979;4:329–337. doi: 10.1111/j.1365-3032.1979.tb00624.x. [DOI] [Google Scholar]
- 85.Dickinson M.H., Tu M.S. The Function of Dipteran Flight Muscle. Comp. Biochem. Physiol. A. 1997;116:223–238. doi: 10.1016/S0300-9629(96)00162-4. [DOI] [Google Scholar]
- 86.Rideout E.J., Billeter J.-C., Goodwin S.F. The Sex-Determination Genes fruitless and doublesex Specify a Neural Substrate Required for Courtship Song. Curr. Biol. 2007;17:1473–1478. doi: 10.1016/j.cub.2007.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rideout E.J., Dornan A.J., Neville M.C., Eadie S., Goodwin S.F. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.von Philipsborn A.C., Shohat-Ophir G., Rezaval C. Single-Pair Courtship and Competition Assays in Drosophila. Cold Spring Harb. Protoc. 2023;2023 doi: 10.1101/pdb.prot108105. pdb.prot108105. [DOI] [PubMed] [Google Scholar]
- 89.von Philipsborn A.C., Shohat-Ophir G., Rezaval C. Courtship Conditioning/Suppression Assays in Drosophila. Cold Spring Harb. Protoc. 2023;2023 doi: 10.1101/pdb.prot108106. pdb.prot108106. [DOI] [PubMed] [Google Scholar]
- 90.von Philipsborn A.C., Shohat-Ophir G., Rezaval C. Probing Acoustic Communication during Fly Reproductive Behaviors. Cold Spring Harb. Protoc. 2023;2023 doi: 10.1101/pdb.prot108107. pdb.prot108107. [DOI] [PubMed] [Google Scholar]
- 91.Arthur B.J., Sunayama-Morita T., Coen P., Murthy M., Stern D.L. Multi-channel acoustic recording and automated analysis of Drosophila courtship songs. BMC Biol. 2013;11:11. doi: 10.1186/1741-7007-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kerwin P., Yuan J., von Philipsborn A.C. Female copulation song is modulated by seminal fluid. Nat. Commun. 2020;11:1430. doi: 10.1038/s41467-020-15260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data is available and will be shared upon request to the lead contact.
This paper did not generate original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.