FIGURE 5.
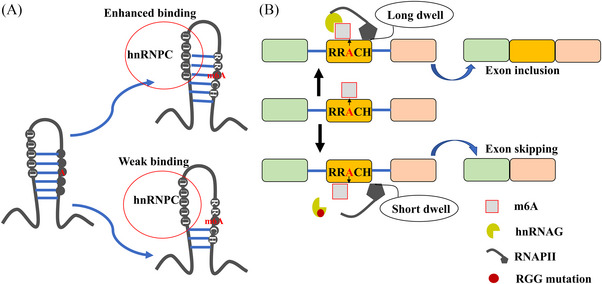
The mechanisms of hnRNPC and hnRNPG in regulating alternative splicing. (A) The m6A modification modulates the stability of the secondary structure of RNAs and facilitates the binding of the reader hnRNPC protein. m6A deposition at the hairpin‐stem of lncRNAs alters the structure stability and contributes to the interaction between hnRNPC and UUUUU motifs, which leads to s alternative splicing of the target RNAs. (B) The RRM and Arg‐Gly‐Gl (RGG) motifs of hnRNPG binds to m6A‐modified RNAs. In addition, it interacts with the phosphorylated C‐terminal domain of RNAP polymerase II (RNAPII) to facilitate its dwell at the exon–intron junction. This dwell increases the splice site utilisation, ultimately inducing exon inclusion. Conversely, mutations in the RGG motifs of hnRNPG induces an opposite effect.
