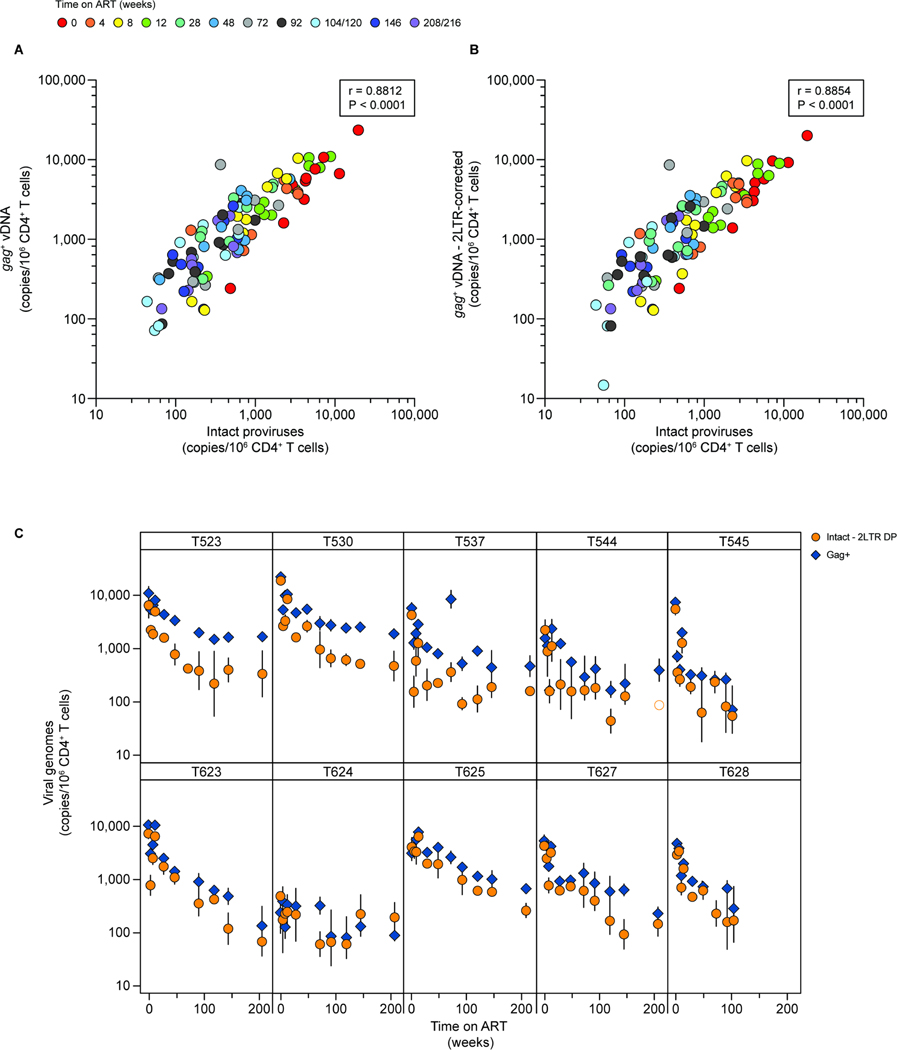
Figure 4. Comparison of gag ddPCR and IPDA quantification of SIV DNA.
(A) Correlation between gag+ SIV DNA copies and intact proviruses quantified using the IPDA. Individual datapoints represent the number of copies detected using either an amplicon in gag (y-axis)67 or the IPDA (x-axis)20. Each point is the geometric mean of 3 replicates. Intact proviruses are corrected for shearing (DSI) and env+2LTR circles. The correlation between the two variables was calculated using Pearson’s coefficient. Data from one animal, T624, were excluded from this analysis due to failure of the gag amplicon resulting from mutations or deletion. (B) Correlation between gag+ SIV DNA copies and intact proviruses, with gag values corrected using the same env+2LTR correction factor applied to the IPDA data, calculated using Pearson’s coefficient. (C) Comparison of the decay of SIV gag copies and intact proviruses for the animals in cohort 18–02. IPDA data are corrected for env+2LTR circles and DNA shearing. Vertical lines represent the standard deviations. See also Figure S4.