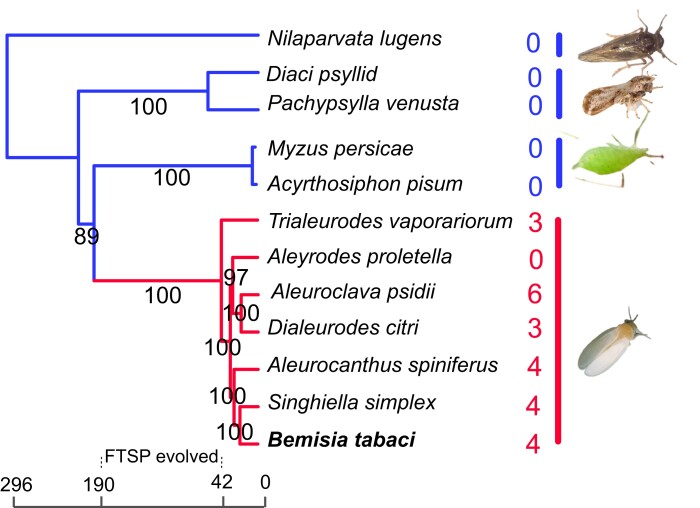
Fig. 1.
Distribution of FTSPs in Hemipteran insects. The coding regions of Trialeurodes vaporariorum, Aleyrodes proletella, Dialeurodes citri, Aleuroclava psidii, Aleurocanthus spiniferus, and Singhiella simplex were predicted using TransDecoder v5.5.0. The predicated proteins from Bemisia tabaci, Acyrthosiphon pisum, Myzus persicae, Diaci psyllid, Pachypsylla venusta, and Nilaparvata lugens were retrieved from a public database. The phylogenetic tree was constructed based on 18 single copy genes. N. lugens was used to root the tree. The divergent time of D. citri- N. lugens (177 to 401 Mya) was used for calibration time estimation. The estimated species divergence time is illustrated at the bottom of the phylogenetic tree. The number of FTSP-related contigs for each species is indicated on the right.