Extended Data Fig. 7 |. Characterization of flexible loops in TnsC.
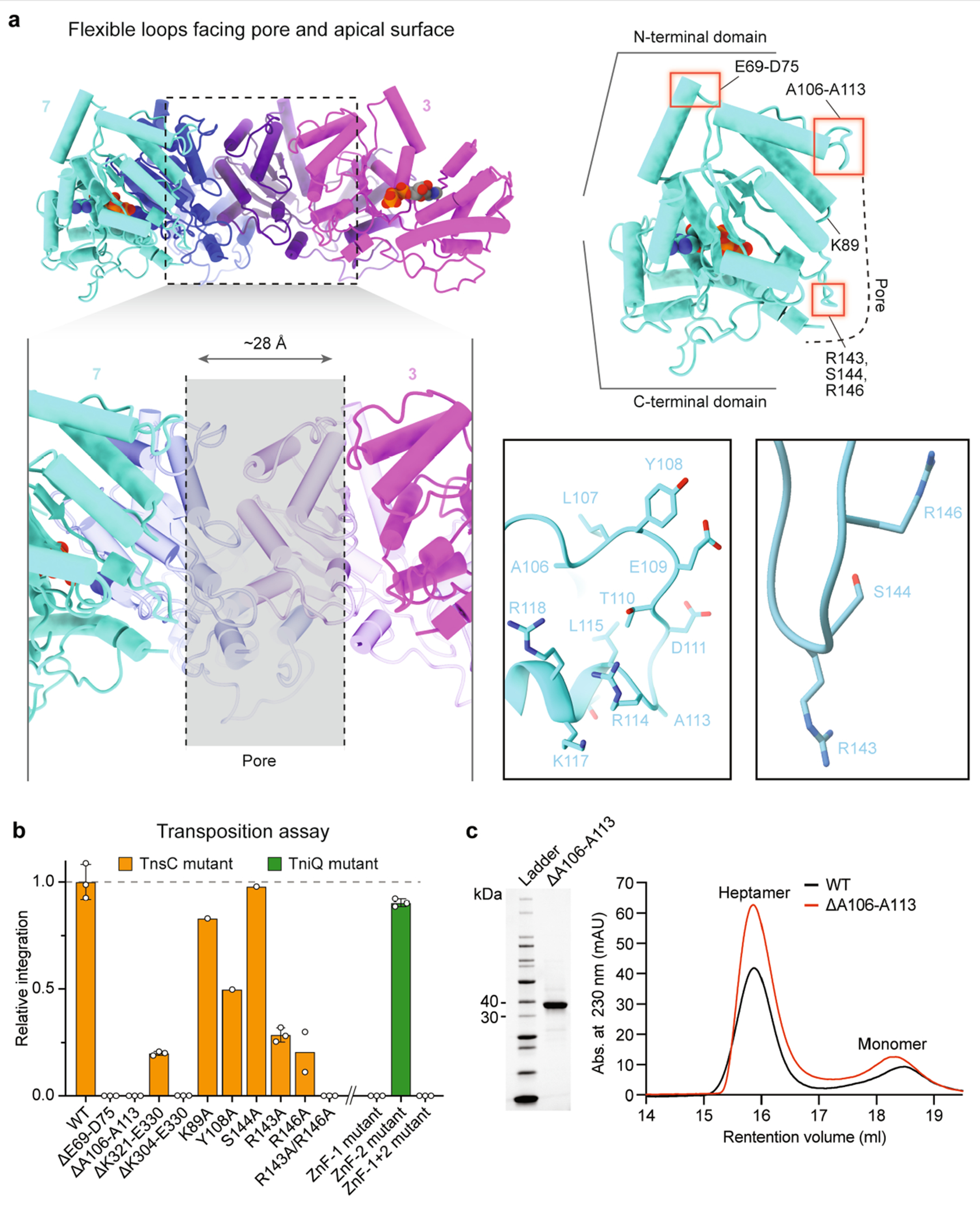
a, Multiple flexible loops within the NTD protrude from the top of the heptameric TnsC disc (top left), which are potential sites of engagement with the TniQ–Cascade complex. Apical loops E69-D75 and A106–A113 are highlighted by red boxes, as are putative DNA-interacting residues including R143, S144 and R146 (top right). A long flexible tail at the CTD folds back towards the centre of the pore, and is likely to engage TnsAB, based on functional similarity to EcoTnsC. Magnified views of A106–A113 and the putative DNA binding region are shown in ribbon representation (centre right), b, CRISPR RNA-guided transposition assays for the indicated TnsC loop mutants, CTD deletions and pore-facing residues (left; coloured in orange), and TniQ ZnF mutations (right; coloured in green), as measured by qPCR. Data are normalized relative to WT. Mutation of R143 and R146 together abrogates transposition. Data are shown as mean ± s.d. of n = 3 independent biological replicates, with the exception of R146A (n = 2) and K89A, Y108A and S144A (n = 1). c, Purification and heptamer assembly experiments with the TnsC A106–A113 loop deletion mutant. SDS-PAGE analysis (left), and SEC chromatograms (right) are shown, with TnsC monomer and TnsC•ATP heptamer peaks indicated. Despite abrogating transposition, this loop deletion mutant is still able to form ATP-dependent heptamers. For TnsC(ΔA106–A113), the baseline was set to 0 at the 5 ml retention volume mark, for clarity. For gel source data, see Supplementary Fig. 1.
