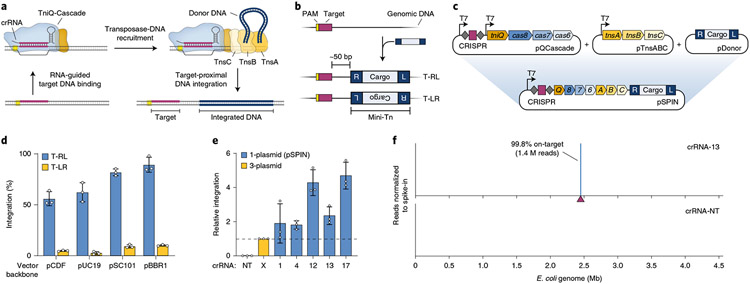
Fig. 1 ∣. Streamlined single-plasmid system for RNA-guided DNA integration.
a, Schematic of INTEGRATE using a Type I-F V. cholerae CRISPR-transposon. b, RNA-guided DNA integration occurs ~50 bp downstream of the target site, in one of two possible orientations (T-RL and T-LR); the mini-transposon (mini-Tn) comprises a genetic cargo flanked by left (L) and right (R) transposon ends. c, Top, a three-plasmid INTEGRATE system encodes protein–RNA components on pQCascade and pTnsABC and the mini-Tn on pDonor. Bottom, a single-plasmid INTEGRATE system (pSPIN) drives protein–RNA expression with a single promoter, on the same vector as the donor DNA. d, qPCR-based quantification of integration efficiency with crRNA-4 for pSPIN with distinct vector backbones and differing copy numbers. e, Relative integration efficiencies for the three-plasmid or single-plasmid (pSPIN) expression system across a non-targeting (NT) and five distinct targeting crRNAs. Data are normalized to the three-plasmid system; pSPIN contained the pBBR1 backbone. f, Normalized Tn-seq data with crRNA-13 and a non-targeting crRNA (crRNA-NT) for pSPIN containing the pBBR1 backbone. Genome-mapping reads are normalized to the reads from a spike-in control (Methods); the target site is denoted by a maroon triangle. Data in d and e are shown as mean ± s.d. for n = 3 biologically independent samples.