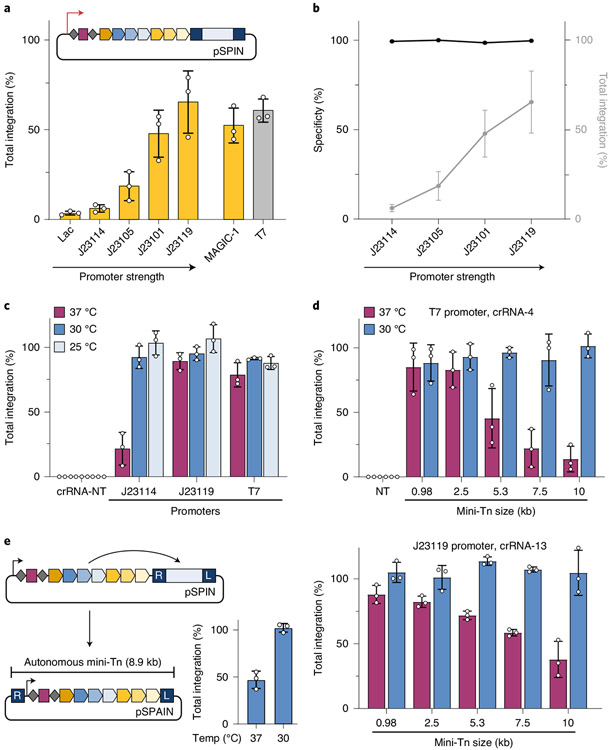
Fig. 2 ∣. INTEGRATE supports high-efficiency insertion of large (10-kb) genetic payloads.
a, qPCR-based quantification of integration efficiency with crRNA-4 as a function of pSPIN promoter identity; MAGiC-1 is taken from ref. 43. b, DNA integration specificity (black) for the promoters shown, as determined by Tn-seq, calculated as the percent of on-target reads relative to all genome-mapping reads (Methods); total integration efficiencies (qPCR) are plotted in gray. c, qPCR-based quantification of integration efficiency with crRNA-4 as a function of culture temperature and promoter strength. Integration reaches ~100% efficiency at lower growth temperatures for all constructs, including the weak J23114 promoter. d, qPCR-based quantification of integration efficiency with variable mini-Tn sizes, after culturing at either 30 °C or 37 °C. The promoter and crRNA used in each panel are shown at the top; experiments were performed with a two-plasmid system comprising pEffector-B (Supplementary Fig. 1c) and pDonor. Unless specified, transposition assays elsewhere in this study use a 0.98-kb mini-Tn. e, Schematic of a single-plasmid autonomous INTEGRATE system (pSPAIN; left) and qPCR-based quantification of integration efficiency with crRNA-4 after culturing at 30 °C and 37 °C (right). The inserted DNA encodes all the necessary machinery for further mobilization. Integration efficiency data in a–e are shown as mean ± s.d. for n = 3 biologically independent samples.