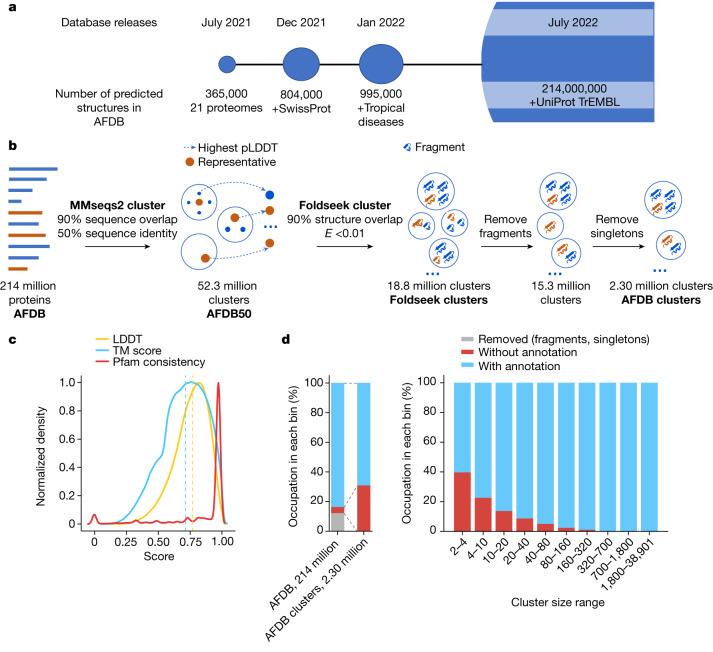
Fig. 1. The AFDB, structural clustering workflow and summary of the clusters.
a, The AFDB started as a collaborative effort between EMBL-EBI and DeepMind in 2021. The database grew in multiple stages, with the latest version of 2022 containing over 214 million predicted protein structures and their confidence metrics. b, A two-step approach was used to cluster proteins in the database. First, MMseqs2 was used to cluster 214 million UniProtKB protein sequences (AFDB) on the basis of 50% sequence identity and 90% sequence overlap, resulting in a reduction of the database size to 52 million clusters (AFDB50). For each cluster, the protein with the highest pLDDT score was selected as the representative. Next, using Foldseek, the representative structures were clustered into 18.8 million clusters (Foldseek clusters) without a sequence identity threshold, but still enforcing a 90% sequence overlap and an E-value of less than 0.01 for each structural alignment. As the last step, we removed all sequences labelled as fragments from the clustering, ending up with 2.30 million clusters with at least two structures (AFDB clusters). c, AFDB cluster structural and Pfam consistency. Our clusters have a median LDDT of 0.77 and a median TM score of 0.71 across all clusters and 66.5% of clusters with Pfam annotations are 100% consistent. d, Summary of sequences and clusters with and without annotation (left) and the relationship of cluster sizes to annotation (right). From left to right, each bin occupies AFDB clusters at rates of 12.24%, 10.59%, 9.20%, 10.07%, 10.46%, 10.05%, 9.04%, 9.20%, 9.19% and 9.96%.