Fig. 2. Putative novel enzymes and small-molecule-binding proteins in structures lacking annotation.
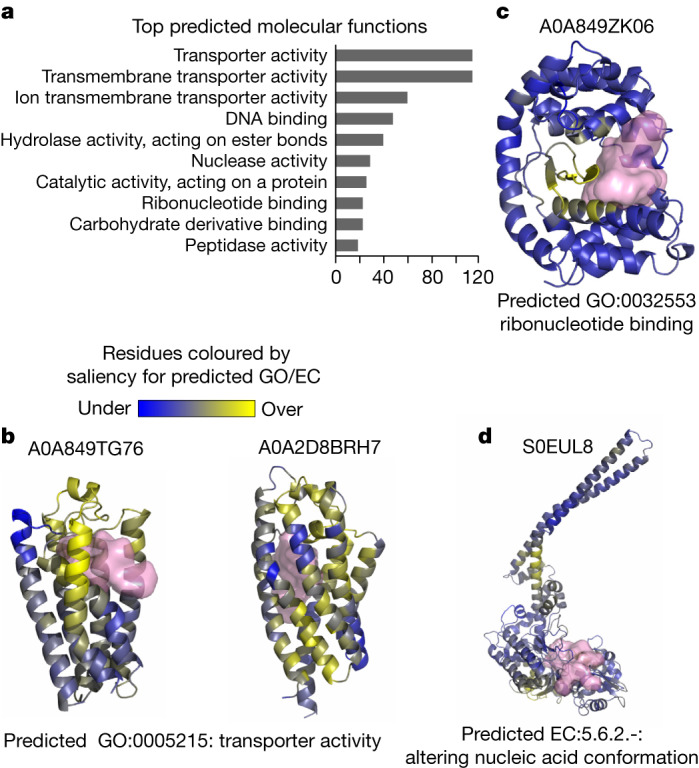
a, Counts of GO molecular function terms that are most often predicted by DeepFRI on the set of selected 1,707 structures with predicted pockets. b–d, Examples of structures (A0A849TG76 and A0A2D8BRH7 (b), A0A849ZK06 (c) and S0EUL8 (d)) with predicted pockets and functional annotations. Each example shows the UniProt ID (top), the highest-scoring DeepFRI function prediction (bottom) and the top-scoring pocket (pink surface). The structures are coloured by residue-level contributions to the DeepFRI function predictions, ranging from blue (no contribution) to yellow (strong contribution).
